Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content
- PMID: 15563614
- PMCID: PMC535842
- DOI: 10.1104/pp.104.053835
Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content
Abstract
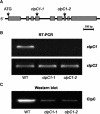
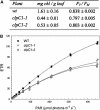
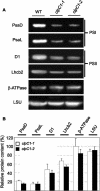
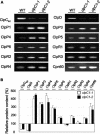
ClpC is a molecular chaperone of the Hsp100 family. In higher plants there are two chloroplast-localized paralogs (ClpC1 and ClpC2) that are approximately 93% similar in primary sequence. In this study, we have characterized two independent Arabidopsis (Arabidopsis thaliana) clpC1 T-DNA insertion mutants lacking on average 65% of total ClpC content. Both mutants display a retarded-growth phenotype, leaves with a homogenous chlorotic appearance throughout all developmental stages, and more perpendicular secondary influorescences. Photosynthetic performance was also impaired in both knockout lines, with relatively fewer photosystem I and photosystem II complexes, but no changes in ATPase and Rubisco content. However, despite the specific drop in photosystem I and photosystem II content, no changes in leaf cell anatomy or chloroplast ultrastructure were observed in the mutants compared to the wild type. Previously proposed functions for envelope-associated ClpC in chloroplast protein import and degradation of mistargeted precursors were examined and shown not to be significantly impaired in the clpC1 mutants. In the stroma, where the majority of ClpC protein is localized, marked increases of all ClpP paralogs were observed in the clpC1 mutants but less variation for the ClpR paralogs and a corresponding decrease in the other chloroplast-localized Hsp100 protein, ClpD. Increased amounts of other stromal molecular chaperones (Cpn60, Hsp70, and Hsp90) and several RNA-binding proteins were also observed. Our data suggest that overall ClpC as a stromal molecular chaperone plays a vital role in chloroplast function and leaf development and is likely involved in photosystem biogenesis.
Figures


Similar articles
-
Mutations in ClpC2/Hsp100 suppress the requirement for FtsH in thylakoid membrane biogenesis.Proc Natl Acad Sci U S A. 2004 Aug 24;101(34):12765-70. doi: 10.1073/pnas.0402764101. Epub 2004 Aug 10. Proc Natl Acad Sci U S A. 2004. PMID: 15304652 Free PMC article.
-
In Vivo Trapping of Proteins Interacting with the Chloroplast CLPC1 Chaperone: Potential Substrates and Adaptors.J Proteome Res. 2019 Jun 7;18(6):2585-2600. doi: 10.1021/acs.jproteome.9b00112. Epub 2019 May 22. J Proteome Res. 2019. PMID: 31070379
-
Quantitative analysis of the chloroplast molecular chaperone ClpC/Hsp93 in Arabidopsis reveals new insights into its localization, interaction with the Clp proteolytic core, and functional importance.J Biol Chem. 2014 Apr 18;289(16):11318-11330. doi: 10.1074/jbc.M113.534552. Epub 2014 Mar 5. J Biol Chem. 2014. PMID: 24599948 Free PMC article.
-
Molecular chaperone involvement in chloroplast protein import.Biochim Biophys Acta. 2013 Feb;1833(2):332-40. doi: 10.1016/j.bbamcr.2012.03.019. Epub 2012 Apr 12. Biochim Biophys Acta. 2013. PMID: 22521451 Review.
-
Arabidopsis variegation mutants: new insights into chloroplast biogenesis.J Exp Bot. 2006;57(9):1871-81. doi: 10.1093/jxb/erj008. Epub 2006 Jan 31. J Exp Bot. 2006. PMID: 16449381 Review.
Cited by
-
A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants.Plant Physiol. 2013 Aug;162(4):1867-80. doi: 10.1104/pp.113.217604. Epub 2013 Jun 26. Plant Physiol. 2013. PMID: 23803583 Free PMC article.
-
Understanding chloroplast biogenesis using second-site suppressors of immutans and var2.Photosynth Res. 2013 Oct;116(2-3):437-53. doi: 10.1007/s11120-013-9855-9. Epub 2013 May 24. Photosynth Res. 2013. PMID: 23703455 Review.
-
ClpC1, an ATP-dependent Clp protease in plastids, is involved in iron homeostasis in Arabidopsis leaves.Ann Bot. 2010 May;105(5):823-33. doi: 10.1093/aob/mcq051. Epub 2010 Apr 9. Ann Bot. 2010. PMID: 20382967 Free PMC article.
-
Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts.Plant Cell. 2010 May;22(5):1516-31. doi: 10.1105/tpc.109.071415. Epub 2010 May 18. Plant Cell. 2010. PMID: 20484004 Free PMC article.
-
Perturbation of protein homeostasis brings plastids at the crossroad between repair and dismantling.PLoS Genet. 2023 Jul 7;19(7):e1010344. doi: 10.1371/journal.pgen.1010344. eCollection 2023 Jul. PLoS Genet. 2023. PMID: 37418499 Free PMC article.
References
-
- Adam Z, Clarke AK (2002) Cutting edge of chloroplast proteolysis. Trends Plant Sci 7: 451–456 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Altman T, Damm B, Halfter U, Willmitzer L, Morris P-C (1992) Protoplast transformation and methods to create specific mutants in Arabidopsis thaliana. In C Koncz, N-H Chua, J Schell, eds, Methods in Arabidopsis Research. World Scientific, London, pp 310–330
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

