Single-track sequencing for genotyping of multiple SNPs in the N-acetyltransferase 1 (NAT1) gene
- PMID: 15563733
- PMCID: PMC544357
- DOI: 10.1186/1472-6750-4-28
Single-track sequencing for genotyping of multiple SNPs in the N-acetyltransferase 1 (NAT1) gene
Abstract
Background: Fast, cheap and reliable methods are needed to identify large populations, which may be at risk in relation to environmental exposure. Polymorphisms in NAT1 (N-acetyl transferase) may be suitable markers to identify individuals at risk.
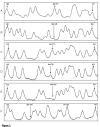
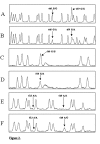
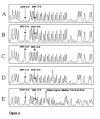
Results: A strategy allowing to address simultaneously 24 various genetic variants in the NAT1 gene using the single sequencing reaction method on the same PCR product is described. A modified automated DNA sequencing using only one of the sequence terminators was used to genotype PCR products in single-track sequencing reactions of NAT1 and was shown to be universal for both DNA sequencing using labeled primers and labeled nucleotides. By this method we detected known SNPs at site T640G, which confers the NAT1*11 allele with frequency of 0.036, further T1088A and C1095A with frequency of 0.172 and 0.188, respectively and a deletion of TAATAATAA in the poly A signal area with a frequency 0.031. All observed frequencies were in Hardy Weinberg equilibrium and comparable to those in Caucasian population. The single-track signatures of the variant genotypes were verified on samples previously genotyped by RLFP.
Conclusions: The method could be of great help to scientists in the field of molecular epidemiology of screening of large populations for known informative biomarkers of susceptibility, such as NAT1.
Figures

References
-
- Kristensen T, Nedelcheva Kristensen V, Børresen-Dale A-L. High throughput screening for known mutations by automated analysis of single sequencing reactions (SSR) BioTechniques. 1998;24:832–835. - PubMed
-
- Kristensen T, Nedelcheva Kristensen V, Børresen-Dale A-L. In "Polymorphism Detection and Analysis". Eaton Publishing; 2000. High-throughput screening for known mutations by automated analysis of single sequencing reactions. - PubMed
-
- Hein DW, Doll MA, Rustan TD, Gray K, Feng Y, Ferguson RJ, Grant DM. Metabolic activation and deactivation of arylamine carcinogens by recombinant human NAT1 and polymorphic NAT2 acetyltransferases. Carcinogenesis. 1993;14:1633–1638. - PubMed
-
- Probst-Hensch NM, Bell DA, Watson MA, Skipper PL, Tannenbaum SR, Chan KK. N-acetyltransferase 2 phenotype but not NAT1*10 genotype affects aminobiphenyl-hemoglobin adduct levels. Cancer Epidemiol. Cancer Epidemiol Biomarkers Prev. 2000;9:619–623. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

