Replicase-binding sites on plus- and minus-strand brome mosaic virus RNAs and their roles in RNA replication in plant cells
- PMID: 15564452
- PMCID: PMC533945
- DOI: 10.1128/JVI.78.24.13420-13429.2004
Replicase-binding sites on plus- and minus-strand brome mosaic virus RNAs and their roles in RNA replication in plant cells
Abstract
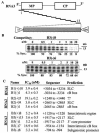
The cis-acting elements for Brome mosaic virus (BMV) RNA synthesis have been characterized primarily for RNA3. To identify additional replicase-binding elements, nested fragments of all three of the BMV RNAs, both plus- and minus-sense fragments, were constructed and tested for binding enriched BMV replicase in a template competition assay. Ten RNA fragments containing replicase-binding sites were identified; eight were characterized further because they were more effective competitors. All eight mapped to noncoding regions of BMV RNAs, and the positions of seven localized to sequences containing previously characterized core promoter elements (C. C. Kao, Mol. Plant Pathol. 3:55-62, 2001), thus suggesting the identities of the replicase-binding sites. Three contained the tRNA-like structures that direct minus-strand RNA synthesis, three were within the 3' region of each minus-strand RNA that contained the core promoter for genomic plus-strand initiation, and one was in the core subgenomic promoter. Single-nucleotide mutations known previously to abolish RNA synthesis in vitro prevented replicase binding. When tested in the context of the respective full-length RNAs, the same mutations abolished BMV RNA synthesis in transfected barley protoplasts. The eighth site was within the intercistronic region (ICR) of plus-strand RNA3. Further mapping showed that a sequence of 22 consecutive adenylates was responsible for binding the replicase, with 16 being the minimal required length. Deletion of the poly(A) sequence was previously shown to severely debilitate BMV RNA replication in plants (E. Smirnyagina, Y. H. Hsu, N. Chua, and P. Ahlquist, Virology 198:427-436, 1994). Interestingly, the B box motif in the ICR of RNA3, which has previously been determined to bind the 1a protein, does not bind the replicase. These results identify the replicase-binding sites in all of the BMV RNAs and suggest that the recognition of RNA3 is different from that of RNA1 and RNA2.
Figures






Similar articles
-
Requirements for brome mosaic virus subgenomic RNA synthesis in vivo and replicase-core promoter interactions in vitro.J Virol. 2004 Jun;78(12):6091-101. doi: 10.1128/JVI.78.12.6091-6101.2004. J Virol. 2004. PMID: 15163702 Free PMC article.
-
Brome mosaic virus RNA syntheses in vitro and in barley protoplasts.J Virol. 2003 May;77(10):5703-11. doi: 10.1128/jvi.77.10.5703-5711.2003. J Virol. 2003. PMID: 12719563 Free PMC article.
-
Template sequence near the initiation nucleotide can modulate brome mosaic virus RNA accumulation in plant protoplasts.J Virol. 2004 Feb;78(3):1169-80. doi: 10.1128/jvi.78.3.1169-1180.2004. J Virol. 2004. PMID: 14722272 Free PMC article.
-
The coat protein leads the way: an update on basic and applied studies with the Brome mosaic virus coat protein.Mol Plant Pathol. 2011 May;12(4):403-12. doi: 10.1111/j.1364-3703.2010.00678.x. Epub 2010 Nov 25. Mol Plant Pathol. 2011. PMID: 21453435 Free PMC article. Review.
-
Common replication strategies emerging from the study of diverse groups of positive-strand RNA viruses.Arch Virol Suppl. 1994;9:181-94. doi: 10.1007/978-3-7091-9326-6_18. Arch Virol Suppl. 1994. PMID: 8032249 Review.
Cited by
-
Characterization of a novel 5' subgenomic RNA3a derived from RNA3 of Brome mosaic bromovirus.J Virol. 2006 Dec;80(24):12357-66. doi: 10.1128/JVI.01207-06. Epub 2006 Sep 27. J Virol. 2006. PMID: 17005659 Free PMC article.
-
Interaction between Brome mosaic virus proteins and RNAs: effects on RNA replication, protein expression, and RNA stability.J Virol. 2005 Nov;79(22):14222-34. doi: 10.1128/JVI.79.22.14222-14234.2005. J Virol. 2005. PMID: 16254357 Free PMC article.
-
Repair of the tRNA-like CCA sequence in a multipartite positive-strand RNA virus.J Virol. 2005 Feb;79(3):1417-27. doi: 10.1128/JVI.79.3.1417-1427.2005. J Virol. 2005. PMID: 15650168 Free PMC article.
-
Phosphorylation of the Brome Mosaic Virus Capsid Regulates the Timing of Viral Infection.J Virol. 2016 Aug 12;90(17):7748-60. doi: 10.1128/JVI.00833-16. Print 2016 Sep 1. J Virol. 2016. PMID: 27334588 Free PMC article.
-
Recombinant viral RdRps can initiate RNA synthesis from circular templates.RNA. 2006 Feb;12(2):303-12. doi: 10.1261/rna.2163106. Epub 2005 Dec 22. RNA. 2006. PMID: 16373481 Free PMC article.
References
-
- Adkins, S., and C. C. Kao. 1998. Subgenomic RNA promoters dictate the mode of recognition by bromoviral RNA-dependent RNA polymerases. Virology 252:1-8. - PubMed
-
- Ahlquist, P. 1992. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 2:271-276. - PubMed
-
- Barrera, I., D. Schuppli, J. M. Sogo, and H. Weber. 1993. Different mechanisms of recognition of bacteriophage Qβ plus and minus strand RNAs by Qβ replicase. J. Mol. Biol. 232:512-521. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

