Assembly protein precursor (pUL80.5 homolog) of simian cytomegalovirus is phosphorylated at a glycogen synthase kinase 3 site and its downstream "priming" site: phosphorylation affects interactions of protein with itself and with major capsid protein
- PMID: 15564461
- PMCID: PMC533919
- DOI: 10.1128/JVI.78.24.13501-13511.2004
Assembly protein precursor (pUL80.5 homolog) of simian cytomegalovirus is phosphorylated at a glycogen synthase kinase 3 site and its downstream "priming" site: phosphorylation affects interactions of protein with itself and with major capsid protein
Abstract
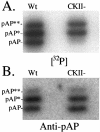
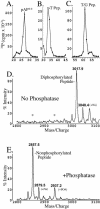
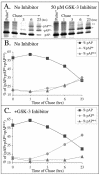
Capsid assembly among the herpes-group viruses is coordinated by two related scaffolding proteins. In cytomegalovirus (CMV), the main scaffolding constituent is called the assembly protein precursor (pAP). Like its homologs in other herpesviruses, pAP is modified by proteolytic cleavage and phosphorylation. Cleavage is essential for capsid maturation and production of infectious virus, but the role of phosphorylation is undetermined. As a first step in evaluating the significance of this modification, we have identified the specific sites of phosphorylation in the simian CMV pAP. Two were established previously to be adjacent serines (Ser156 and Ser157) in a casein kinase II consensus sequence. The remaining two, identified here as Thr231 and Ser235, are within consensus sequences for glycogen synthase kinase 3 (GSK-3) and mitogen-activated protein kinase, respectively. Consistent with Thr231 being a GSK-3 substrate, its phosphorylation required a downstream "priming" phosphate (i.e., Ser235) and was reduced by a GSK-3-specific inhibitor. Phosphorylation of Ser235 converts pAP to an electrophoretically slower-mobility isoform, pAP*; subsequent phosphorylation of pAP* at Thr231 converts pAP* to a still-slower isoform, pAP**. The mobility shift to pAP* was mimicked by substituting an acidic amino acid for either Thr231 or Ser235, but the shift to pAP** required that both positions be phosphorylated. Glu did not substitute for pSer235 in promoting phosphorylation of Thr231. We suggest that phosphorylation of Thr231 and Ser235 causes charge-driven conformational changes in pAP, and we demonstrate that preventing these modifications alters interactions of pAP with itself and with major capsid protein, suggesting a functional significance.
Figures








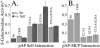

References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
-
- Coghlan, M. P., A. A. Culbert, D. A. Cross, S. L. Corcoran, J. W. Yates, N. J. Pearce, O. L. Rausch, G. J. Murphy, P. S. Carter, L. R. Cox, D. Mills, M. J. Brown, D. Haigh, R. W. Ward, D. G. Smith, K. J. Murray, A. D. Reith, and J. C. Holder. 2000. Selective small molecule inhibitors of glycogen synthase kinase-3 modulate glycogen metabolism and gene transcription. Chem. Biol. 7:793-803. - PubMed
-
- Desai, P., and S. Person. 1996. Molecular interactions between the HSV-1 capsid proteins as measured by the yeast two-hybrid system. Virology 220:516-521. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

