Antiviral effects of human immunodeficiency virus type 1-specific small interfering RNAs against targets conserved in select neurotropic viral strains
- PMID: 15564478
- PMCID: PMC533941
- DOI: 10.1128/JVI.78.24.13687-13696.2004
Antiviral effects of human immunodeficiency virus type 1-specific small interfering RNAs against targets conserved in select neurotropic viral strains
Abstract
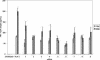
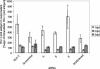
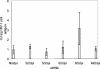
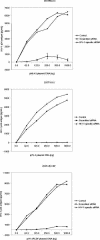
RNA interference, a natural biological phenomenon mediated by small interfering RNAs (siRNAs), has been demonstrated in recent studies to be an effective strategy against human immunodeficiency virus type 1 (HIV-1). In the present study, we used 21-bp chemically synthesized siRNA duplexes whose sequences were derived from the gp41 gene, nef, tat, and rev regions of viral RNA. These sequences are conserved in select neurotropic strains of HIV-1 (JR-FL, JR-CSF, and YU-2). The designed siRNAs exerted a potent antiviral effect on these HIV-1 strains. The antiviral effect was mediated at the RNA level (as observed by the down-regulation of the HIV-1-specific spliced transcript generating a 1.2-kbp reverse transcription [RT]-PCR product) as well as viral assembly on the cell membrane. Spliced transcripts (apart from the most abundant transcript generating a 1.2-kbp RT-PCR product) arising from an unspliced precursor likely contributed, albeit to a lesser extent, to the antiviral effect. The resultant progeny viruses had infectivities similar to that of input virus. We therefore conclude that these siRNAs interfere with the processing of the unspliced transcripts for the gp41 gene, tat, rev, and nef, eventually affecting viral assembly and leading to the overall inhibition of viral production. Apart from using the gp41 gene as a target, the conservation of each of these targets in the above-mentioned viral strains, as well as several primary isolates, would enable these siRNAs to be used as potent antiviral tools for investigations with cells derived from the central nervous system in order to evaluate their therapeutic potential and assess their utility in inhibiting HIV-1 neuropathogenesis and neuroinvasion.
Figures



Similar articles
-
Temporal patterns of human immunodeficiency virus type 1 transcripts in human fetal astrocytes.J Virol. 1994 Jan;68(1):93-102. doi: 10.1128/JVI.68.1.93-102.1994. J Virol. 1994. PMID: 8254781 Free PMC article.
-
Down-modulation of TCR/CD3 surface complexes after HIV-1 infection is associated with differential expression of the viral regulatory genes.Eur J Immunol. 2001 Apr;31(4):969-79. doi: 10.1002/1521-4141(200104)31:4<969::aid-immu969>3.0.co;2-2. Eur J Immunol. 2001. PMID: 11298321
-
Splicing regulatory elements within tat exon 2 of human immunodeficiency virus type 1 (HIV-1) are characteristic of group M but not group O HIV-1 strains.J Virol. 1999 Dec;73(12):9764-72. doi: 10.1128/JVI.73.12.9764-9772.1999. J Virol. 1999. PMID: 10559286 Free PMC article.
-
The tat gene and protein of the human immunodeficiency virus type 1.New Microbiol. 1995 Jan;18(1):87-110. New Microbiol. 1995. PMID: 7760763 Review. No abstract available.
-
HIV-1 inactivation by nucleic acid aptamers.Front Biosci. 2006 Jan 1;11:89-112. doi: 10.2741/1782. Front Biosci. 2006. PMID: 16146716 Review.
Cited by
-
Lentviral-mediated RNAi to inhibit target gene expression of the porcine integrin αv subunit, the FMDV receptor, and against FMDV infection in PK-15 cells.Virol J. 2011 Sep 7;8:428. doi: 10.1186/1743-422X-8-428. Virol J. 2011. PMID: 21899738 Free PMC article.
-
Developing neuroprotective strategies for treatment of HIV-associated neurocognitive dysfunction.Futur HIV Ther. 2008;2(3):271-280. doi: 10.2217/17469600.2.3.271. Futur HIV Ther. 2008. PMID: 19774095 Free PMC article.
-
Morphine affects HIV-induced inflammatory response without influencing viral replication in human monocyte-derived macrophages.FEMS Immunol Med Microbiol. 2012 Mar;64(2):228-36. doi: 10.1111/j.1574-695X.2011.00894.x. FEMS Immunol Med Microbiol. 2012. PMID: 22066570 Free PMC article.
-
Transgenic shRNA pigs reduce susceptibility to foot and mouth disease virus infection.Elife. 2015 Jun 19;4:e06951. doi: 10.7554/eLife.06951. Elife. 2015. PMID: 26090904 Free PMC article.
-
Interfering antiviral immunity: application, subversion, hope?Trends Immunol. 2006 Jul;27(7):328-35. doi: 10.1016/j.it.2006.05.006. Epub 2006 Jun 6. Trends Immunol. 2006. PMID: 16753342 Free PMC article. Review.
References
-
- Abbas-Terki, T., W. Blanco-Bose, N. Deglon, W. Pralong, and P. Aebischer. 2002. Lentiviral-mediated RNA interference. Hum. Gene Ther. 13:2197-2201. - PubMed
-
- Anderson, J., A. Banerjea, V. Planelles, and R. Akkina. 2003. Potent suppression of HIV type 1 infection by a short hairpin anti-CXCR4 siRNA. AIDS Res. Hum. Retrovir. 19:699-706. - PubMed
-
- Aquaro, S., R. Calio, J. Balzarini, M. C. Bellocchi, E. Garaci, and C. F. Perno. 2002. Macrophages and HIV infection: therapeutical approaches toward this strategic virus reservoir. Antiviral Res. 55:209-225. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

