Fitness costs limit viral escape from cytotoxic T lymphocytes at a structurally constrained epitope
- PMID: 15564498
- PMCID: PMC533946
- DOI: 10.1128/JVI.78.24.13901-13910.2004
Fitness costs limit viral escape from cytotoxic T lymphocytes at a structurally constrained epitope
Abstract
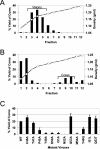
The intense selection pressure exerted by virus-specific cytotoxic T lymphocytes (CTL) on replicating human immunodeficiency virus and simian immunodeficiency virus results in the accumulation of CTL epitope mutations. It has been assumed that fitness costs can limit the evolution of CTL epitope mutations. However, only a limited number of studies have carefully examined this possibility. To explore the fitness costs associated with viral escape from p11C, C-M-specific CTL, we constructed a panel of viruses encoding point mutations at each position of the entire p11C, C-M epitope. Amino acid substitutions at positions 3, 4, 5, 6, 7, and 9 of the epitope significantly impaired virus replication by altering virus production and Gag protein expression as well as by destabilizing mature cores. Amino acid substitutions at position 2 of the epitope were tolerated but required reversion or additional compensatory mutations to generate replication-competent viruses. Finally, while amino acid substitutions at positions 1 and 8 of the p11C, C-M epitope were functionally tolerated, these substitutions were recognized by p11C, C-M-specific CTL and therefore provided no selection advantage for the virus. Together, these data suggest that limited sequence variation is tolerated by the region of the capsid encoding the p11C, C-M epitope and therefore that only a very limited number of mutations can allow successful viral escape from the p11C, C-M-specific CTL response.
Figures




Similar articles
-
Compensatory substitutions restore normal core assembly in simian immunodeficiency virus isolates with Gag epitope cytotoxic T-lymphocyte escape mutations.J Virol. 2006 Aug;80(16):8168-77. doi: 10.1128/JVI.00068-06. J Virol. 2006. PMID: 16873273 Free PMC article.
-
Simian-human immunodeficiency virus escape from cytotoxic T-lymphocyte recognition at a structurally constrained epitope.J Virol. 2003 Dec;77(23):12572-8. doi: 10.1128/jvi.77.23.12572-12578.2003. J Virol. 2003. PMID: 14610180 Free PMC article.
-
Cytotoxic T lymphocytes do not appear to select for mutations in an immunodominant epitope of simian immunodeficiency virus gag.J Immunol. 1992 Dec 15;149(12):4060-6. J Immunol. 1992. PMID: 1460291
-
Vaccination reduces simian-human immunodeficiency virus sequence reversion through enhanced viral control.J Virol. 2010 Dec;84(24):12782-9. doi: 10.1128/JVI.01193-10. Epub 2010 Sep 29. J Virol. 2010. PMID: 20881040 Free PMC article.
-
Cytotoxic T-lymphocyte escape viral variants: how important are they in viral evasion of immune clearance in vivo?Immunol Rev. 1998 Aug;164(1):37-51. doi: 10.1111/j.1600-065x.1998.tb01206.x. Immunol Rev. 1998. PMID: 9795762 Free PMC article. Review.
Cited by
-
Low level of HIV-1 evolution after transmission from mother to child.Sci Rep. 2014 May 28;4:5079. doi: 10.1038/srep05079. Sci Rep. 2014. PMID: 24866155 Free PMC article.
-
Stable cytotoxic T cell escape mutation in hepatitis C virus is linked to maintenance of viral fitness.PLoS Pathog. 2008 Sep 5;4(9):e1000143. doi: 10.1371/journal.ppat.1000143. PLoS Pathog. 2008. PMID: 18773115 Free PMC article.
-
HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC.PLoS Pathog. 2009 May;5(5):e1000414. doi: 10.1371/journal.ppat.1000414. Epub 2009 May 8. PLoS Pathog. 2009. PMID: 19424423 Free PMC article.
-
Evolutionary gamut of in vivo Gag substitutions during early HIV-1 subtype C infection.Virology. 2011 Dec 20;421(2):119-28. doi: 10.1016/j.virol.2011.09.020. Epub 2011 Oct 19. Virology. 2011. PMID: 22014506 Free PMC article.
-
Compensatory substitutions restore normal core assembly in simian immunodeficiency virus isolates with Gag epitope cytotoxic T-lymphocyte escape mutations.J Virol. 2006 Aug;80(16):8168-77. doi: 10.1128/JVI.00068-06. J Virol. 2006. PMID: 16873273 Free PMC article.
References
-
- Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. - PubMed
-
- Barouch, D. H., J. Kunstman, J. Glowczwskie, K. J. Kunstman, M. A. Egan, F. W. Peyerl, S. Santra, M. J. Kuroda, J. E. Schmitz, K. Beaudry, G. R. Krivulka, M. A. Lifton, D. A. Gorgone, S. M. Wolinsky, and N. L. Letvin. 2003. Viral escape from dominant simian immunodeficiency virus epitope-specific cytotoxic T lymphocytes in DNA-vaccinated rhesus monkeys. J. Virol. 77:7367-7375. - PMC - PubMed
-
- Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. - PubMed
-
- Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

