Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha
- PMID: 15565168
- PMCID: PMC544904
- DOI: 10.1038/sj.emboj.7600491
Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha
Abstract
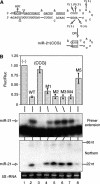
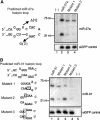
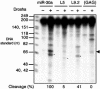
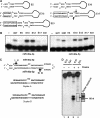
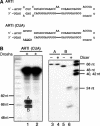
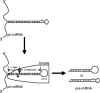
A critical step during human microRNA maturation is the processing of the primary microRNA transcript by the nuclear RNaseIII enzyme Drosha to generate the approximately 60-nucleotide precursor microRNA hairpin. How Drosha recognizes primary RNA substrates and selects its cleavage sites has remained a mystery, especially given that the known targets for Drosha processing show no discernable sequence homology. Here, we show that human Drosha selectively cleaves RNA hairpins bearing a large (>/=10 nucleotides) terminal loop. From the junction of the loop and the adjacent stem, Drosha then cleaves approximately two helical RNA turns into the stem to produce the precursor microRNA. Beyond the precursor microRNA cleavage sites, approximately one helix turn of stem extension is also essential for efficient processing. While the sites of Drosha cleavage are determined largely by the distance from the terminal loop, variations in stem structure and sequence around the cleavage site can fine-tune the actual cleavage sites chosen.
Figures



References
-
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 - PubMed
-
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNA in mammalian cells. Science 296: 550–553 - PubMed
-
- Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301: 336–338 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

