Cloning of CviPII nicking and modification system from chlorella virus NYs-1 and application of Nt.CviPII in random DNA amplification
- PMID: 15570069
- PMCID: PMC535667
- DOI: 10.1093/nar/gkh958
Cloning of CviPII nicking and modification system from chlorella virus NYs-1 and application of Nt.CviPII in random DNA amplification
Abstract
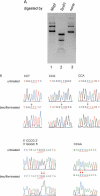
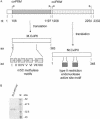
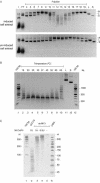
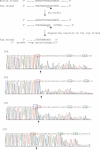
The cloning and expression of the CviPII DNA nicking and modification system encoded by chlorella virus NYs-1 is described. The system consists of a co-linear MTase encoding gene (cviPIIM) and a nicking endonuclease encoding gene (cviPIINt) separated by 12 nt. M.CviPII possesses eight conserved amino acid motifs (I to VIII) typical of C5 MTases, but, like another chlorella virus MTase M.CviJI, lacks conserved motifs IX and X. In addition to modification of the first cytosine in CCD (D = A, G or T) sequences, M.CviPII modifies both the first two cytosines in CCAA and CCCG sites as well. Nt.CviPII has significant amino acid sequence similarity to Type II restriction endonuclease CviJI that recognizes an overlapping sequence (RG--CY). Nt.CviPII was expressed in Escherichia coli with or without a His-tag in a host pre-modified by M.CviPII. Recombinant Nt.CviPII recognizes the DNA sequence CCD and cleaves the phosphodiester bond 5' of the first cytosine while the other strand of DNA at this site is not affected. Nt.CviPII displays site preferences with CCR (R = A or G) sites preferred over CCT sites. Nt.CviPII is active from 16 to 65 degrees C with a temperature optimum of 30-45 degrees C. Nt.CviPII can be used to generate single-stranded DNAs (ssDNAs) for isothermal strand-displacement amplification. Nt.CviPII was used in combination with Bst DNA polymerase I large fragment to rapidly amplify anonymous DNA from genomic DNA or from a single bacterial colony.
Figures



Similar articles
-
Cloning of Nt.CviQII nicking endonuclease and its cognate methyltransferase: M.CviQII methylates AG sequences.Protein Expr Purif. 2006 Sep;49(1):138-50. doi: 10.1016/j.pep.2006.04.002. Epub 2006 Apr 25. Protein Expr Purif. 2006. PMID: 16737828
-
A single amino acid change restores DNA cytosine methyltransferase activity in a cloned chlorella virus pseudogene.Nucleic Acids Res. 1992 Apr 11;20(7):1637-42. doi: 10.1093/nar/20.7.1637. Nucleic Acids Res. 1992. PMID: 1579454 Free PMC article.
-
Molecular cloning of the three base restriction endonuclease R.CviJI from eukaryotic Chlorella virus IL-3A.Nucleic Acids Res. 1996 Jul 1;24(13):2463-9. doi: 10.1093/nar/24.13.2463. Nucleic Acids Res. 1996. PMID: 8692682 Free PMC article.
-
DNA methyltransferases and DNA site-specific endonucleases encoded by chlorella viruses.EXS. 1993;64:186-211. doi: 10.1007/978-3-0348-9118-9_9. EXS. 1993. PMID: 8380349 Review. No abstract available.
-
Natural and engineered nicking endonucleases--from cleavage mechanism to engineering of strand-specificity.Nucleic Acids Res. 2011 Jan;39(1):1-18. doi: 10.1093/nar/gkq742. Epub 2010 Aug 30. Nucleic Acids Res. 2011. PMID: 20805246 Free PMC article. Review.
Cited by
-
Cofactor requirement of HpyAV restriction endonuclease.PLoS One. 2010 Feb 5;5(2):e9071. doi: 10.1371/journal.pone.0009071. PLoS One. 2010. PMID: 20140205 Free PMC article.
-
Engineering variants of the I-SceI homing endonuclease with strand-specific and site-specific DNA-nicking activity.J Mol Biol. 2008 Sep 26;382(1):188-202. doi: 10.1016/j.jmb.2008.07.010. Epub 2008 Jul 11. J Mol Biol. 2008. PMID: 18644379 Free PMC article.
-
Site- and strand-specific nicking of DNA by fusion proteins derived from MutH and I-SceI or TALE repeats.Nucleic Acids Res. 2013 Apr;41(7):e83. doi: 10.1093/nar/gkt080. Epub 2013 Feb 13. Nucleic Acids Res. 2013. PMID: 23408850 Free PMC article.
-
Engineering nicking enzymes that preferentially nick 5-methylcytosine-modified DNA.Nucleic Acids Res. 2014 May;42(9):e77. doi: 10.1093/nar/gku192. Epub 2014 Mar 7. Nucleic Acids Res. 2014. PMID: 24609382 Free PMC article.
-
Chimeric DNA byproducts in strand displacement amplification using the T7 replisome.PLoS One. 2022 Sep 19;17(9):e0273979. doi: 10.1371/journal.pone.0273979. eCollection 2022. PLoS One. 2022. PMID: 36121810 Free PMC article.
References
-
- Morgan R.D., Calvet,C., Demeter,M., Agra,R. and Kong,H. (2000) Characterization of the specific DNA nicking activity of restriction endonuclease. N. BstNBI. Biol. Chem., 381, 1123–1125. - PubMed
-
- Abdurashitov M.A., Belichenko,O.A., Shevchenko,A.V. and Degtyarev,S.K. (1996) N.BstSE—site-specific nuclease from Bacillus stearothermophilus SE-589—restriction endonuclease production. Molek. Biol., 30, 1261–1267. - PubMed
-
- Zhang Y., Nelson,M., Nietfeldt,J., Xia,Y., Burbank,D., Ropp,S. and Van Etten,J.L. (1998) Chlorella virus NY-2A encodes at least 12 DNA endonuclease/methyltransferase genes. Virology, 240, 366–375. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

