A simulation model of Escherichia coli osmoregulatory switch using E-CELL system
- PMID: 15571621
- PMCID: PMC543474
- DOI: 10.1186/1471-2180-4-44
A simulation model of Escherichia coli osmoregulatory switch using E-CELL system
Abstract
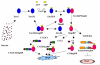
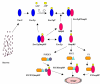
Background: Bacterial signal transduction mechanism referred to as a "two component regulatory systems" contributes to the overall adaptability of the bacteria by regulating the gene expression. Osmoregulation is one of the well-studied two component regulatory systems comprising of the sensor, EnvZ and the cognate response regulator, OmpR, which together control the expression of OmpC and OmpF porins in response to the osmolyte concentration.
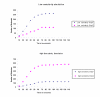
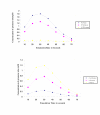
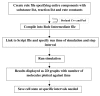
Results: A quantitative model of the osmoregulatory switch operative in Escherichia coli was constructed by integrating the enzyme rate equations using E-CELL system. Using the substance reactor logic of the E-CELL system, a total of 28 reactions were defined from the injection of osmolyte till the regulated expression of porins by employing the experimental kinetic constants as reported in literature. In the case of low osmolarity, steady state production of OmpF and repression of OmpC was significant. In this model we show that the steady state - production of OmpF is dramatically reduced in the high osmolarity medium. The rate of OmpC production increased after sucrose addition, which is comparable with literature results. The relative porin production seems to be unaltered with changes in cell volume changes, ATP, EnvZ and OmpR at low and high osmolarity conditions. But the reach of saturation was rapid at high and low osmolarity with altered levels of the above components.
Conclusions: The E-CELL system allows us to perform virtual experiments on the bacterial osmoregulation model. This model does not take into account interaction with other networks in the cell. It suggests that the regulation of OmpF and OmpC is a direct consequence of the level of OmpRP in the cell and is dependent on the way in which OmpRP interacts with ompF and ompC regulatory regions. The preliminary simulation experiment indicates that both reaching steady state expression and saturation is delayed in the case of OmpC compared to OmpF. Experimental analysis will help improve the model. The model captures the basic features of the generally accepted view of EnvZ-OmpR signaling and is a reasonable starting point for building sophisticated models and explaining quantitative features of the system.
Figures

References
-
- Takahashi K, Yugi K, Hashimoto K, Yamada Y, Pickett C, Tomita M. Computational challenges in cell simulation a software Engineering Approach. IEEE Intelligent Systems. 2002;17:64–71. doi: 10.1109/MIS.2002.1039834. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

