Structural basis for peptidoglycan binding by peptidoglycan recognition proteins
- PMID: 15572450
- PMCID: PMC535381
- DOI: 10.1073/pnas.0407856101
Structural basis for peptidoglycan binding by peptidoglycan recognition proteins
Abstract
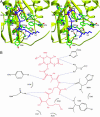
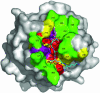
Peptidoglycan (PGN) recognition proteins (PGRPs) are pattern-recognition receptors of the innate immune system that bind and, in some cases, hydrolyze bacterial PGNs. We determined the crystal structure, at 2.30-A resolution, of the C-terminal PGN-binding domain of human PGRP-Ialpha in complex with a muramyl tripeptide representing the core of lysine-type PGNs from Gram-positive bacteria. The peptide stem of the ligand is buried at the deep end of a long binding groove, with N-acetylmuramic acid situated in the middle of the groove, whose shallow end can accommodate a linked N-acetylglucosamine. Although most interactions are with the peptide, the glycan moiety also seems to be essential for specific recognition by PGRPs. Conservation of key PGN-contacting residues shows that all PGRPs employ this basic PGN-binding mode. The structure pinpoints variable residues that likely mediate discrimination between lysine- and diaminopimelic acid-type PGNs. We also propose a mechanism for PGN hydrolysis by Zn(2+)-containing PGRPs.
Figures
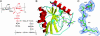
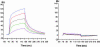
References
-
- Medzhitov, R. & Janeway, C. A., Jr. (2002) Science 296, 298–300. - PubMed
-
- Hoffmann, J. A. (2003) Nature 426, 33–38. - PubMed
-
- Weis, W. I., Taylor, M. E. & Drickamer, K. (1998) Immunol. Rev. 163, 19–34. - PubMed
-
- van Heijenoort, J. (2001) Glycobiology 11, 25R–36R. - PubMed
-
- Doyle, R. J. & Dziarski, R. (2001) in Molecular Medical Microbiology, ed. Sussman, M. (Academic, London), pp. 137–153.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

