Three-dimensional structure and organization of a receptor/signaling complex
- PMID: 15572451
- PMCID: PMC536031
- DOI: 10.1073/pnas.0407826101
Three-dimensional structure and organization of a receptor/signaling complex
Abstract
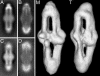
Transmembrane signaling in bacterial chemotaxis has become an important model system for experimental and theoretical studies. These studies have provided a wealth of detailed molecular structures, including the structures of CheA, CheW, and the cytoplasmic domain of the serine receptor Tsr. How these three proteins interact to form the receptor/signaling complex remains unknown. By using EM and single-particle image analysis, we present a three-dimensional reconstruction of the receptor/signaling complex. The complex contains CheA, CheW, and the cytoplasmic portion of the aspartate receptor Tar. We observe density consistent with a structure containing 24 aspartate-receptor monomers and additional density sufficient to house the expected four CheA monomers and six CheW monomers. Within this bipolar structure are four groups of three receptor dimers that are not threefold symmetric and are therefore unlike the symmetric trimers observed in the x-ray crystal structure of the cytoplasmic domain of the serine receptor. In the latter, the interdimer contacts occur in the signaling domains near the hairpin loop. In our structure, the signaling domains within trimers appear spaced apart by the presence of CheA and CheW. This structure argues against models where one CheA and one CheW bind to the outer face of each of the dimers in the trimer. This structure of the receptor/signaling complex provides an additional basis for understanding the architecture of the large arrays of chemotaxis receptors, CheA, and CheW found at the cell poles in motile bacteria.
Figures



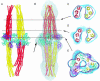

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

