Reconstructing large regions of an ancestral mammalian genome in silico
- PMID: 15574820
- PMCID: PMC534665
- DOI: 10.1101/gr.2800104
Reconstructing large regions of an ancestral mammalian genome in silico
Erratum in
- Genome Res. 2005 Mar;15(3):451
Abstract
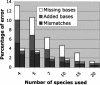
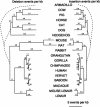
It is believed that most modern mammalian lineages arose from a series of rapid speciation events near the Cretaceous-Tertiary boundary. It is shown that such a phylogeny makes the common ancestral genome sequence an ideal target for reconstruction. Simulations suggest that with methods currently available, we can expect to get 98% of the bases correct in reconstructing megabase-scale euchromatic regions of an eutherian ancestral genome from the genomes of approximately 20 optimally chosen modern mammals. Using actual genomic sequences from 19 extant mammals, we reconstruct 1.1 Mb of ancient genome sequence around the CFTR locus. Detailed examination suggests the reconstruction is accurate and that it allows us to identify features in modern species, such as remnants of ancient transposon insertions, that were not identified by direct analysis. Tracing the predicted evolutionary history of the bases in the reconstructed region, estimates are made of the amount of DNA turnover due to insertion, deletion, and substitution in the different placental mammalian lineages since the common eutherian ancestor, showing considerable variation between lineages. In coming years, such reconstructions may help in identifying and understanding the genetic features common to eutherian mammals and may shed light on the evolution of human or primate-specific traits.
Figures

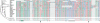
References
-
- Bejerano, G., Pheasant, M., Makunin, I., Stephen, S., Kent, W.J., Mattick, J.S., and Haussler, D. 2004. Ultraconserved elements in the human genome. Science 304: 1321-1325. - PubMed
-
- Bienvenu, T., Petitpretz, P., Beldjord, C., and Kaplan, J.C. 1994. A missense mutation (F87L) in exon 3 of the cystic fibrosis transmembrane conductance regulator gene. Hum. Mutat. 3: 395-396. - PubMed
Web site references
-
- www.nisc.nih.gov; NISC Comparative Sequencing Program.
-
- http://genome.ucsc.edu/ancestors; Author's Supplemental information site.
-
- http://genome.ucsc.edu; Interactive browser for alignments.
-
- http://www.genet.sickkids.on.ca/cftr/; Cystic Fibrosis Mutation Database.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
