Characterization of the novel Fusobacterium nucleatum plasmid pKH9 and evidence of an addiction system
- PMID: 15574887
- PMCID: PMC535169
- DOI: 10.1128/AEM.70.12.6957-6962.2004
Characterization of the novel Fusobacterium nucleatum plasmid pKH9 and evidence of an addiction system
Abstract
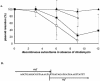
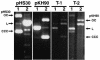
Fusobacterium nucleatum is an important oral anaerobic pathogen involved in periodontal and systemic infections. Studies of the molecular mechanisms involved in fusobacterial virulence and adhesion have been limited by lack of systems for efficient genetic manipulation. Plasmids were isolated from eight strains of F. nucleatum. The smallest plasmid, pKH9 (4,975 bp), was characterized and used to create new vectors for fusobacterial genetic manipulation. DNA sequence analysis of pKH9 revealed an open reading frame (ORF) encoding a putative autonomous rolling circle replication protein (Rep), an ORF predicted to encode a protein homologous to members of the FtsK/SpoIIIE cell division-DNA segregation protein family, and an operon encoding a putative toxin-antitoxin plasmid addiction system (txf-axf). Deletion analysis localized the pKH9 replication region in a 0.96-kbp fragment. The pKH9 rep gene is not present in this fragment, suggesting that pKH9 can replicate in fusobacteria independently of the Rep protein. A pKH9-based, compact Escherichia coli-F. nucleatum shuttle plasmid was constructed and found to be compatible with a previously described pFN1-based fusobacterial shuttle plasmid. Deletion of the pKH9 putative addiction system (txf-axf) reduced plasmid stability in fusobacteria, indicating its addiction properties and suggesting it to be the first plasmid addiction system described for fusobacteria. pKH9, its genetic elements, and its shuttle plasmid derivatives can serve as useful tools for investigating fusobacterial properties important in biofilm ecology and pathogenesis.
Figures


References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Bachrach, G., G. Rosen, M. Bellalou, and M. Sela. 2004. Identification of a Fusobacterium nucleatum 65 kDa serine protease. Oral Microbiol. Immunol. 19:155-159. - PubMed
-
- Bath, J., L. J. Wu, J. Errington, and J. C. Wang. 2000. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science 290:995-997. - PubMed
-
- Christensen, S. K., G. Maenhaut-Michel, N. Mine, S. Gottesman, K. Gerdes, and L. Van Melderen. 2004. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol. Microbiol. 51:1705-1717. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

