Use of Pseudomonas putida EstA as an anchoring motif for display of a periplasmic enzyme on the surface of Escherichia coli
- PMID: 15574889
- PMCID: PMC535197
- DOI: 10.1128/AEM.70.12.6968-6976.2004
Use of Pseudomonas putida EstA as an anchoring motif for display of a periplasmic enzyme on the surface of Escherichia coli
Abstract
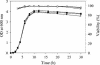
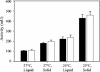
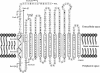
The functional expression of proteins on the surface of bacteria has proven important for numerous biotechnological applications. In this report, we investigated the N-terminal fusion display of the periplasmic enzyme beta-lactamase (Bla) on the surface of Escherichia coli by using the translocator domain of the Pseudomonas putida outer membrane esterase (EstA), which is a member of the lipolytic autotransporter enzymes. To find out the transport function of a C-terminal domain of EstA, we generated a set of Bla-EstA fusion proteins containing N-terminally truncated derivatives of the EstA C-terminal domain. The surface exposure of the Bla moiety was verified by whole-cell immunoblots, protease accessibility, and fluorescence-activated cell sorting. The investigation of growth kinetics and host cell viability showed that the presence of the EstA translocator domain in the outer membrane neither inhibits cell growth nor affects cell viability. Furthermore, the surface-exposed Bla moiety was shown to be enzymatically active. These results demonstrate for the first time that the translocator domain of a lipolytic autotransporter enzyme is an effective anchoring motif for the functional display of heterologous passenger protein on the surface of E. coli. This investigation also provides a possible topological model of the EstA translocator domain, which might serve as a basis for the construction of fusion proteins containing heterologous passenger domains.
Figures




Similar articles
-
Probing the applicability of autotransporter based surface display with the EstA autotransporter of Pseudomonas stutzeri A15.Microb Cell Fact. 2012 Dec 13;11:158. doi: 10.1186/1475-2859-11-158. Microb Cell Fact. 2012. PMID: 23237539 Free PMC article.
-
A generic system for the Escherichia coli cell-surface display of lipolytic enzymes.FEBS Lett. 2005 Feb 14;579(5):1177-82. doi: 10.1016/j.febslet.2004.12.087. FEBS Lett. 2005. PMID: 15710409
-
Functional display of Pseudomonas and Burkholderia lipases using a translocator domain of EstA autotransporter on the cell surface of Escherichia coli.J Biotechnol. 2010 Apr 1;146(3):126-9. doi: 10.1016/j.jbiotec.2010.01.022. Epub 2010 Feb 6. J Biotechnol. 2010. PMID: 20138931
-
Autotransporters with GDSL passenger domains: molecular physiology and biotechnological applications.Chembiochem. 2011 Jul 4;12(10):1476-85. doi: 10.1002/cbic.201100013. Epub 2011 May 19. Chembiochem. 2011. PMID: 21598370 Review.
-
The expression of recombinant proteins on the external surface of Escherichia coli. Biotechnological applications.Ann N Y Acad Sci. 1994 Nov 30;745:372-82. doi: 10.1111/j.1749-6632.1994.tb44389.x. Ann N Y Acad Sci. 1994. PMID: 7832524 Review.
Cited by
-
Bacterial display using circularly permuted outer membrane protein OmpX yields high affinity peptide ligands.Protein Sci. 2006 Apr;15(4):825-36. doi: 10.1110/ps.051897806. Protein Sci. 2006. PMID: 16600968 Free PMC article.
-
Of linkers and autochaperones: an unambiguous nomenclature to identify common and uncommon themes for autotransporter secretion.Mol Microbiol. 2015 Jan;95(1):1-16. doi: 10.1111/mmi.12838. Epub 2014 Nov 24. Mol Microbiol. 2015. PMID: 25345653 Free PMC article. Review.
-
Probing the applicability of autotransporter based surface display with the EstA autotransporter of Pseudomonas stutzeri A15.Microb Cell Fact. 2012 Dec 13;11:158. doi: 10.1186/1475-2859-11-158. Microb Cell Fact. 2012. PMID: 23237539 Free PMC article.
-
Surface display of proteins by gram-negative bacterial autotransporters.Microb Cell Fact. 2006 Jun 20;5:22. doi: 10.1186/1475-2859-5-22. Microb Cell Fact. 2006. PMID: 16787545 Free PMC article.
-
Functional cell surface display and controlled secretion of diverse Agarolytic enzymes by Escherichia coli with a novel ligation-independent cloning vector based on the autotransporter YfaL.Appl Environ Microbiol. 2012 May;78(9):3051-8. doi: 10.1128/AEM.07004-11. Epub 2012 Feb 17. Appl Environ Microbiol. 2012. PMID: 22344647 Free PMC article.
References
-
- Agterberg, M., H. Adriaanse, A. van Bruggen, M. Karperien, and J. Tommassen. 1990. Outer-membrane PhoE protein of Escherichia coli K-12 as an exposure vector: possibilities and limitations. Gene 88:37-45. - PubMed
-
- Benhar, I. 2001. Biotechnological applications of phage and cell display. Biotechnol. Adv. 19:1-33. - PubMed
-
- Bingle, W. H., J. F. Nomellini, and J. Smit. 1997. Cell-surface display of a Pseudomonas aeruginosa strain K pilin peptide within the paracrystalline S-layer of Caulobacter crescentus. Mol. Microbiol. 26:277-288. - PubMed
-
- Bolivar, F. 1978. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique EcoRI sites for selection of EcoRI generated recombinant DNA molecules. Gene 4:121-136. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

