Microarray and functional gene analyses of sulfate-reducing prokaryotes in low-sulfate, acidic fens reveal cooccurrence of recognized genera and novel lineages
- PMID: 15574893
- PMCID: PMC535148
- DOI: 10.1128/AEM.70.12.6998-7009.2004
Microarray and functional gene analyses of sulfate-reducing prokaryotes in low-sulfate, acidic fens reveal cooccurrence of recognized genera and novel lineages
Abstract
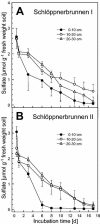
Low-sulfate, acidic (approximately pH 4) fens in the Lehstenbach catchment in the Fichtelgebirge mountains in Germany are unusual habitats for sulfate-reducing prokaryotes (SRPs) that have been postulated to facilitate the retention of sulfur and protons in these ecosystems. Despite the low in situ availability of sulfate (concentration in the soil solution, 20 to 200 microM) and the acidic conditions (soil and soil solution pHs, approximately 4 and 5, respectively), the upper peat layers of the soils from two fens (Schlöppnerbrunnen I and II) of this catchment displayed significant sulfate-reducing capacities. 16S rRNA gene-based oligonucleotide microarray analyses revealed stable diversity patterns for recognized SRPs in the upper 30 cm of both fens. Members of the family "Syntrophobacteraceae" were detected in both fens, while signals specific for the genus Desulfomonile were observed only in soils from Schlöppnerbrunnen I. These results were confirmed and extended by comparative analyses of environmentally retrieved 16S rRNA and dissimilatory (bi)sulfite reductase (dsrAB) gene sequences; dsrAB sequences from Desulfobacca-like SRPs, which were not identified by microarray analysis, were obtained from both fens. Hypotheses concerning the ecophysiological role of these three SRP groups in the fens were formulated based on the known physiological properties of their cultured relatives. In addition to these recognized SRP lineages, six novel dsrAB types that were phylogenetically unrelated to all known SRPs were detected in the fens. These dsrAB sequences had no features indicative of pseudogenes and likely represent novel, deeply branching, sulfate- or sulfite-reducing prokaryotes that are specialized colonists of low-sulfate habitats.
Figures




References
-
- Abdelouas, A., W. Lutze, W. Gong, E. H. Nuttall, B. A. Strietelmeier, and B. J. Travis. 2000. Biological reduction of uranium in groundwater and subsurface soil. Sci. Total Environ. 250:21-35. - PubMed
-
- Adachi, J., and M. Hasegawa. 1996. MOLPHY version 2.3: programs for molecular phylogenetics based on maximum likelihood. Comput. Sci. Monogr. 28:1-150.
-
- Alewell, C., and M. Gehre. 1999. Patterns of stable S isotopes in a forested catchment as indicators for biological S turnover. Biogeochemistry 47:319-333.
-
- Alewell, C., and A. Giesemann. 1996. Sulfate reduction in a forested catchment as indicated by d34S values of sulfate in soil solutions and runoff. Isot. Environ. Health Stud. 32:203-210. - PubMed
-
- Alewell, C., and M. Novak. 2001. Spotting zones of dissimilatory sulfate reduction in a forested catchment: the 34S-35S approach. Environ. Pollut. 112:369-377. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

