Biosynthetic gene cluster of the herbicide phosphinothricin tripeptide from Streptomyces viridochromogenes Tü494
- PMID: 15574905
- PMCID: PMC535184
- DOI: 10.1128/AEM.70.12.7093-7102.2004
Biosynthetic gene cluster of the herbicide phosphinothricin tripeptide from Streptomyces viridochromogenes Tü494
Abstract
The antibiotic phosphinothricin tripeptide (PTT) consists of two molecules of L-alanine and one molecule of the unusual amino acid phosphinothricin (PT) which are nonribosomally combined. The bioactive compound PT has bactericidal, fungicidal, and herbicidal properties and possesses a C-P-C bond, which is very rare in natural compounds. Previously uncharacterized flanking and middle regions of the PTT biosynthetic gene cluster from Streptomyces viridochromogenes Tü494 were isolated and sequenced. The boundaries of the gene cluster were identified by gene inactivation studies. Sequence analysis and homology searches led to the completion of the gene cluster, which consists of 24 genes. Four of these were identified as undescribed genes coding for proteins that are probably involved in uncharacterized early steps of antibiotic biosynthesis or in providing precursors of PTT biosynthesis (phosphoenolpyruvate, acetyl-coenzyme A, or L-alanine). The involvement of the genes orfM and trs and of the regulatory gene prpA in PTT biosynthesis was analyzed by gene inactivation and overexpression, respectively. Insight into the regulation of PTT was gained by determining the transcriptional start sites of the pmi and prpA genes. A previously undescribed regulatory gene involved in morphological differentiation in streptomycetes was identified outside of the left boundary of the PTT biosynthetic gene cluster.
Figures



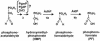
References
-
- Alijah, R., J. Dorendorf, S. Talay, A. Pühler, and W. Wohlleben. 1991. Genetic analysis of the phosphinothricin-tripeptide biosynthetic pathway of Streptomyces viridochromogenes Tü 494. Appl. Microbiol. Biotechnol. 34:749-755. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Bayer, E., K. H. Gugel, K. Hägele, H. Hagenmaier, S. Jassipow, W. A. König, and H. Zähner. 1972. Stoffwechselprodukte von Mikroorganismen. Phosphinothricin und Phosphinothricyl-Alanyl-Alanin. Helv. Chim. Acta 55:224-239. - PubMed
-
- Bibb, M. J., P. R. Findlay, and M. W. Johnson. 1984. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene 30:157-166. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

