Streptomyces coelicolor A3(2) lacks a genomic island present in the chromosome of Streptomyces lividans 66
- PMID: 15574907
- PMCID: PMC535201
- DOI: 10.1128/AEM.70.12.7110-7118.2004
Streptomyces coelicolor A3(2) lacks a genomic island present in the chromosome of Streptomyces lividans 66
Abstract
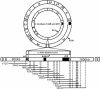
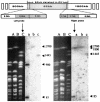
Streptomyces lividans ZX1 has become a preferred host for DNA cloning in Streptomyces species over its progenitor, the wild-type strain 66 (stock number 1326 from the John Innes Center collection), especially when stable DNA is crucial for in vitro electrophoresis, because DNA from strain 66 contains a novel modification that makes it sensitive to oxidative double-strand cleavage during electrophoresis. Detailed analysis of this modification-deficient mutant (ZX1) revealed that it has several additional phenotypic traits associated with a chromosomal deletion of ca. 90 kb, which was cloned and mapped by using a cosmid library. Comparative sequence analysis of two clones containing the left and right deletion ends originating from strain 66 and one clone with the deletion and fused sequence cloned from strain ZX1 revealed a perfect 15-bp direct repeat, which may have mediated deletion and fusion to yield strain ZX1 by site-specific recombination. Analysis of AseI linking clones in the deleted region in relation to the published AseI map of strain ZX1 yielded a complete AseI map for the S. lividans 66 genome, on which the relative positions of a cloned phage phiHAU3 resistance (phiHAU3r) gene and the dnd gene cluster were precisely localized. Comparison of S. lividans ZX1 and its progenitor 66, as well as the sequenced genome of its close relative, Streptomyces coelicolor M145, reveals that the ca. 90-kb deletion in strain ZX1 may have originated from an insertion from an unknown source.
Figures



References
-
- Bao, K., Z. Hu, X. Zhou, Q. Zhou, and Z. Deng. 1997. A bifunctional cosmid vector for the mobilized conjugal transfer of DNA from E. coli to Streptomyces sp. Prog. Nat. Sci. 7:568-572.
-
- Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. - PubMed
-
- Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. - PubMed
-
- Borck, K., J. D. Beggs, W. J. Brammar, A. S. Hopkins, and N. E. Murray. 1976. The construction in vitro of transducing derivatives of phage lambda. Mol. Gen. Genet. 146:199-207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

