Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces
- PMID: 15574920
- PMCID: PMC535136
- DOI: 10.1128/AEM.70.12.7220-7228.2004
Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces
Abstract
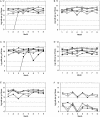
16S rRNA gene-targeted group-specific primers were designed and validated for specific detection and quantification of the Clostridium leptum subgroup and the Atopobium cluster. To monitor the predominant bacteria in human feces by real-time PCR, we used these specific primers together with four sets of group-specific primers for the Clostridium coccoides group, the Bacteroides fragilis group, Bifidobacterium, and Prevotella developed in a previous study (T. Matsuki, K. Watanabe, J. Fujimoto, Y. Miyamoto, T. Takada, K. Matsumoto, H. Oyaizu, and R. Tanaka, Appl. Environ. Microbiol. 68:5445-5451, 2002). Examination of DNA extracted from the feces of 46 healthy adults showed that the C. coccoides group was present in the greatest numbers (log10 10.3 +/- 0.3 cells per g [wet weight] [average +/- standard deviation]), followed by the C. leptum subgroup (log10 9.9 +/- 0.7 cells per g [wet weight]), the B. fragilis group (log10 9.9 +/- 0.3 cells per g [wet weight]), Bifidobacterium (log10 9.4 +/- 0.7 cells per g [wet weight]), and the Atopobium cluster (log10 9.3 +/- 0.7 cells per g [wet weight]). These five bacterial groups were detected in all 46 volunteers. Prevotella was found in only 46% of the subjects at a level of log10 9.7 +/- 0.8 cells per g (wet weight). Examination of changes in the population and the composition of the intestinal flora for six healthy adults over an 8-month period revealed that the composition of the flora of each volunteer remained stable throughout the test period.
Figures
References
-
- Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. - PubMed
-
- Finegold, S. M., H. R. Attebery, and V. L. Sutter. 1974. Effect of diet on human fecal flora: comparison of Japanese and American diets. Am. J. Clin. Nutr. 27:1456-1469. - PubMed
-
- Finegold, S. M., V. S. Sutter, and G. E. Mathisen. 1983. Normal indigenous intestinal flora, p. 3-31. In D. J. Hentges (ed.), Human intestinal microflora in health and disease. Academic Press, New York, N.Y.
-
- Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

