The role of phosphate groups in the VS ribozyme-substrate interaction
- PMID: 15576350
- PMCID: PMC535666
- DOI: 10.1093/nar/gkh957
The role of phosphate groups in the VS ribozyme-substrate interaction
Abstract
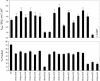
The VS ribozyme trans-cleavage substrate interacts with the catalytic RNA via tertiary interactions. To study the role of phosphate groups in the ribozyme-substrate interaction, 18 modified substrates were synthesized, where an epimeric phosphorothioate replaces one of the phosphate diester linkages. Sites in the stem-loop substrate where phosphorothioate substitution impaired reaction cluster in two regions. The first site is the scissile phosphate diester linkage and nucleotides downstream of this and the second site is within the loop region. The addition of manganese ions caused recovery of the rate of reaction for phosphorothioate substitutions between A621 and A622 and U631 and C632, suggesting that these two phosphate groups may serve as ligands for two metal ions. In contrast, significant manganese rescue was not observed for the scissile phosphate diester linkage implying that electrophilic catalysis by metal ions is unlikely to contribute to VS ribozyme catalysis. In addition, an increase in the reaction rate of the unmodified VS ribozyme was observed when a mixture of magnesium and manganese ions acted as the cofactor. One possible explanation for this effect is that the cleavage reaction of the VS ribozyme is rate limited by a metal dependent docking of the substrate on the ribozyme.
Figures
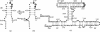
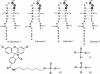


References
-
- Guo H.C.T., Deabreu,D.M., Tillier,E.R.M., Saville,B.J., Olive,J.E. and Collins,R.A. (1993) Nucleotide-sequence requirements for self-cleavage of Neurospora VS RNA. J. Mol. Biol., 232, 351–361. - PubMed
-
- Andersen A.A. and Collins,R.A. (2000) Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol. Cell., 5, 469–478. - PubMed

