DmGEN, a novel RAD2 family endo-exonuclease from Drosophila melanogaster
- PMID: 15576351
- PMCID: PMC535671
- DOI: 10.1093/nar/gkh962
DmGEN, a novel RAD2 family endo-exonuclease from Drosophila melanogaster
Abstract
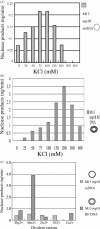
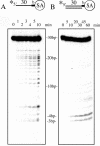
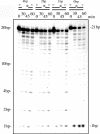
A novel endo-exonuclease, DmGEN (Drosophila Melanogaster XPG-like endonuclease), was identified in D.melanogaster. DmGEN is composed of five exons and four introns, and the open reading frame encodes a predicted product of 726 amino acid residues with a molecular weight of 82.5 kDa and a pI of 5.36. The gene locus on Drosophila polytene chromosomes was detected at 64C9 on the left arm of chromosome 3 as a single site. The encoded protein showed a relatively high degree of sequence homology with the RAD2 nucleases, especially XPG. Although the XPG-N- and XPG-I-domains are highly conserved in sequence, locations of the domains are similar to those of FEN-1 and EXO-1, and the molecular weight of the protein is close to that of EXO-1. In vitro, DmGEN showed endonuclease and 3'-5' exonuclease activities with both single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA), but the endonuclease action with dsDNA was quite specific: 5'-3' exonuclease activity was found to occur with nicked DNA, while dsDNA was endonucleolytically cut at 3-4 bp from the 5' end. Homologs are widely found in mammals and higher plants. The data suggest that DmGEN belongs to a new class of RAD2 nuclease.
Figures






References
-
- Kimura S., Suzuki,T., Yanagawa,Y., Yamamoto,T., Nakagawa,H., Tanaka,I., Hashimoto,J. and Sakaguchi,K. (2001) Characterization of plant proliferating cell nuclear antigen (PCNA) and flap endonuclease-1 (FEN-1), and their distribution in mitotic and meiotic cell cycles. Plant J., 28, 643–653. - PubMed
-
- Kimura S., Ueda,T., Hatanaka,M., Takenouchi,M., Hashimoto,J. and Sakaguchi,K. (2000) Plant homologue of flap endonuclease-1: molecular cloning, characterization, and evidence of expression in meristematic tissues. Plant Mol. Biol., 42, 415–427. - PubMed
-
- Furukawa T., Kimura,S., Ishibashi,T., Mori,Y., Hashimoto,J. and Sakaguchi,K. (2003) OsSEND-1: a new RAD2 nuclease family member in higher plants. Plant Mol. Biol., 51, 59–70. - PubMed
-
- Calleja F.M., Nivard,M.J. and Eeken,J.C. (2001) Induced mutagenic effects in the nucleotide excision repair deficient Drosophila mutant mus201(D1), expressing a truncated XPG protein. Mutat. Res., 461, 279–288. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

