Differences in the folding transition state of ubiquitin indicated by phi and psi analyses
- PMID: 15576508
- PMCID: PMC536030
- DOI: 10.1073/pnas.0407683101
Differences in the folding transition state of ubiquitin indicated by phi and psi analyses
Abstract
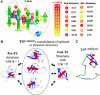
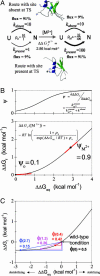
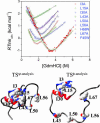
We compare the folding transition state (TS) of ubiquitin previously identified by using psi analysis to that determined by using analysis. Both methods attempt to identify interactions and their relative populations at the rate-limiting step for folding. The TS ensemble derived from psi analysis has an extensive native-like chain topology, with a four-stranded beta-sheet network and a portion of the major helix. According to analysis, however, the TS is much smaller and more polarized, with only a local helix/hairpin motif. We find that structured regions can have values far from unity, the canonical value for such sites, because of structural relaxation of the TS. Consequently, these sites may be incorrectly interpreted as contributing little to the structure of the TS. These results stress the need for caution when interpreting and drawing conclusions from analysis alone and highlight the need for more specific tools for examining the structure and energetics of the TS ensemble.
Figures
Comment in
-
Phi value versus psi analysis.Proc Natl Acad Sci U S A. 2004 Dec 14;101(50):17327-8. doi: 10.1073/pnas.0407863101. Epub 2004 Dec 6. Proc Natl Acad Sci U S A. 2004. PMID: 15583125 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

