Electric charge balance mechanism of extended soluble proteins
- PMID: 15576568
- PMCID: PMC2253322
- DOI: 10.1110/ps.04984505
Electric charge balance mechanism of extended soluble proteins
Abstract
Extended proteins such as calmodulin and troponin C have two globular terminal domains linked by a central region that is exposed to water and often acts as a function-regulating element. The mechanisms that stabilize the tertiary structure of extended proteins appear to differ greatly from those of globular proteins. Identifying such differences in physical properties of amino acid sequences between extended proteins and globular proteins can provide clues useful for identification of extended proteins from complete genomes including orphan sequences. In the present study, we examined the structure and amino acid sequence of extended proteins. We found that extended proteins have a large net electric charge, high charge density, and an even balance of charge between the terminal domains, indicating that electrostatic interaction is a dominant factor in stabilization of extended proteins. Additionally, the central domain exposed to water contained many amphiphilic residues. Extended proteins can be identified from these physical properties of the tertiary structure, which can be deduced from the amino acid sequence. Analysis of physical properties of amino acid sequences can provide clues to the mechanism of protein folding. Also, structural changes in extended proteins may be caused by formation of molecular complexes. Long-range effects of electrostatic interactions also appear to play important roles in structural changes of extended proteins.
Figures
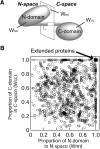



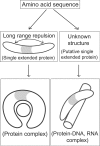
References
-
- Babu, Y.S., Bugg, C.E., and Cook, W.J. 1988. Structure of calmodulin refined at 2.2 Å resolution. J. Mol. Biol. 204 191–204. - PubMed
-
- Chou, J.J., Li, S., Klee, C.B., and Bax, A. 2001. Solution structure of Ca(2+)-calmodulin reveals flexible hand-like properties of its domains. Nat. Struct. Biol. 8 990–997. - PubMed
-
- Hardin, C., Pogorelov, T.V., and Luthey-Schulten, Z. 2002. Ab initio protein structure prediction. Curr. Opin. Struct. Biol. 12 176–181. - PubMed
-
- Hirokawa, T., Boon-Chieng, S., and Mitaku, S. 1998. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics 14 378–379. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

