Comparative sequence analysis of IS50/Tn5 transposase
- PMID: 15576772
- PMCID: PMC532410
- DOI: 10.1128/JB.186.24.8240-8247.2004
Comparative sequence analysis of IS50/Tn5 transposase
Abstract
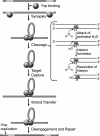
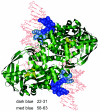
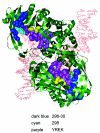
Comparative sequence analysis of IS50 transposase-related protein sequences in conjunction with known structural, biochemical, and genetic data was used to determine domains and residues that play key roles in IS50 transposase function. BLAST and ClustalW analyses have been used to find and analyze six complete protein sequences that are related to the IS50 transposase. The protein sequence identity of these six homologs ranged from 25 to 55% in comparison to the IS50 transposase. Homologous motifs were found associated with each of the three catalytic residues. Residues that play roles in transposase-DNA binding, protein autoregulation, and DNA hairpin formation were also found to be conserved in addition to other residues of unknown function. On the other hand, some homologous sequences did not appear to be competent to encode the inhibitor regulatory protein. The results were also used to compare the IS50 transposase with the more distantly related transposase encoded by IS10.
Figures



Similar articles
-
Defining functional regions of the IS903 transposase.J Mol Biol. 1997 Dec 12;274(4):491-504. doi: 10.1006/jmbi.1997.1410. J Mol Biol. 1997. PMID: 9417930
-
Tn5 transposase loops DNA in the absence of Tn5 transposon end sequences.Mol Microbiol. 2006 Dec;62(6):1558-68. doi: 10.1111/j.1365-2958.2006.05471.x. Mol Microbiol. 2006. PMID: 17074070
-
Presence of a characteristic D-D-E motif in IS1 transposase.J Bacteriol. 2002 Nov;184(22):6146-54. doi: 10.1128/JB.184.22.6146-6154.2002. J Bacteriol. 2002. PMID: 12399484 Free PMC article.
-
Comparative architecture of transposase and integrase complexes.Nat Struct Biol. 2001 May;8(5):302-7. doi: 10.1038/86166. Nat Struct Biol. 2001. PMID: 11774877 Review.
-
Comparative architecture of transposase and integrase complexes.Nat Struct Biol. 2001 Apr;8(4):302-7. doi: 10.1038/86166. Nat Struct Biol. 2001. PMID: 11276247 Review.
Cited by
-
Phage WO of Wolbachia: lambda of the endosymbiont world.Trends Microbiol. 2010 Apr;18(4):173-81. doi: 10.1016/j.tim.2009.12.011. Epub 2010 Jan 18. Trends Microbiol. 2010. PMID: 20083406 Free PMC article. Review.
-
A bifunctional DNA binding region in Tn5 transposase.Mol Microbiol. 2008 Feb;67(3):528-40. doi: 10.1111/j.1365-2958.2007.06056.x. Epub 2007 Dec 14. Mol Microbiol. 2008. PMID: 18086215 Free PMC article.
-
Phosphate coordination and movement of DNA in the Tn5 synaptic complex: role of the (R)YREK motif.Nucleic Acids Res. 2008 Oct;36(18):5855-62. doi: 10.1093/nar/gkn577. Epub 2008 Sep 12. Nucleic Acids Res. 2008. PMID: 18790806 Free PMC article.
-
Tn5 synaptic complex formation: role of transposase residue W450.J Bacteriol. 2008 Feb;190(4):1484-7. doi: 10.1128/JB.01488-07. Epub 2007 Dec 14. J Bacteriol. 2008. PMID: 18083803 Free PMC article.
-
Insertion sequence diversity in archaea.Microbiol Mol Biol Rev. 2007 Mar;71(1):121-57. doi: 10.1128/MMBR.00031-06. Microbiol Mol Biol Rev. 2007. PMID: 17347521 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

