Effect of mild acid on gene expression in Staphylococcus aureus
- PMID: 15576791
- PMCID: PMC532443
- DOI: 10.1128/JB.186.24.8407-8423.2004
Effect of mild acid on gene expression in Staphylococcus aureus
Abstract
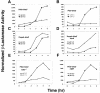
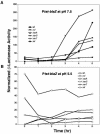
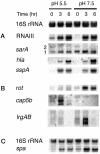
During staphylococcal growth in glucose-supplemented medium, the pH of a culture starting near neutrality typically decreases by about 2 units due to the fermentation of glucose. Many species can comfortably tolerate the resulting mildly acidic conditions (pH, approximately 5.5) by mounting a cellular response, which serves to defend the intracellular pH and, in principle, to modify gene expression for optimal performance in a mildly acidic infection site. In this report, we show that changes in staphylococcal gene expression formerly thought to represent a glucose effect are largely the result of declining pH. We examine the cellular response to mild acid by microarray analysis and define the affected gene set as the mild acid stimulon. Many of the genes encoding extracellular virulence factors are affected, as are genes involved in regulation of virulence factor gene expression, transport of sugars and peptides, intermediary metabolism, and pH homeostasis. Key results are verified by gene fusion and Northern blot hybridization analyses. The results point to, but do not define, possible regulatory pathways by which the organism senses and responds to a pH stimulus.
Figures



References
-
- Arvidson, S., and T. Holme. 1971. Influence of pH on the formation of extracellular proteins by Staphylococcus aureus. Acta Pathol. Microbiol. Scand. B 79:406-413. - PubMed
-
- Arvidson, S., T. Holme, and T. Wadstrom. 1971. Influence of cultivation conditions on the production of extracellular proteins by Staphylococcus aureus. Acta Pathol. Microbiol. Scand. B 79:399-405. - PubMed
-
- Bergdoll, M. S. 1989. Regulation and control of toxic shock syndrome toxin 1: overview. Rev. Infect. Dis. 11(Suppl. 1):S142-S144. - PubMed
-
- Bessman, A. N., J. Page, and L. J. Thomas. 1989. In vivo pH of induced soft-tissue abscesses in diabetic and nondiabetic mice. Diabetes 38:659-662. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

