Specific NEMO mutations impair CD40-mediated c-Rel activation and B cell terminal differentiation
- PMID: 15578091
- PMCID: PMC529497
- DOI: 10.1172/JCI21345
Specific NEMO mutations impair CD40-mediated c-Rel activation and B cell terminal differentiation
Abstract
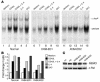
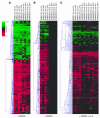
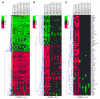
Hypomorphic mutations in the zinc finger domain of NF-kappaB essential modulator (NEMO) cause X-linked hyper-IgM syndrome with ectodermal dysplasia (XHM-ED). Here we report that patient B cells are characterized by an absence of Ig somatic hypermutation (SHM) and defective class switch recombination (CSR) despite normal induction of activation-induced cytidine deaminase (AID) and Iepsilon-Cepsilon transcripts. This indicates that AID expression alone is insufficient to support neutralizing antibody responses. Furthermore, we show that patient B cells stimulated with CD40 ligand are impaired in both p65 and c-Rel activation, and whereas addition of IL-4 can enhance p65 activity, c-Rel activity remains deficient. This suggests that these NF-kappaB components have different activation requirements and that IL-4 can augment some but not all NEMO-dependent NF-kappaB signaling. Finally, using microarray analysis of patient B cells we identified downstream effects of impaired NF-kappaB activation and candidate factors that may be necessary for CSR and SHM in B cells.
Figures

References
-
- Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. - PubMed
-
- Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. - PubMed
-
- Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. - PubMed
-
- Yoshikawa K, et al. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science. 2002;296:2033–2036. - PubMed
-
- Karin M, Ben-Neriah Y. Phos-phorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu. Rev. Immunol. 2000;18:621–663. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

