The amino terminus of the Saccharomyces cerevisiae DNA helicase Rrm3p modulates protein function altering replication and checkpoint activity
- PMID: 15579680
- PMCID: PMC1448792
- DOI: 10.1534/genetics.104.028035
The amino terminus of the Saccharomyces cerevisiae DNA helicase Rrm3p modulates protein function altering replication and checkpoint activity
Abstract
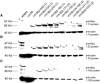
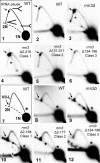
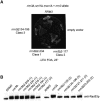
The Pif1 family of DNA helicases is conserved from yeast to humans. Although the helicase domains of family members are well conserved, the amino termini of these proteins are not. The Saccharomyces cerevisiae genome encodes two Pif1 family members, Rrm3p and Pif1p, that have very different functions. To determine if the amino terminus of Rrm3p contributes to its role in promoting fork progression at >1000 discrete chromosomal sites, we constructed a deletion series that lacked portions of the 249-amino-acid amino terminus. The phenotypes of cells expressing alleles that lacked all or most of the amino terminus were indistinguishable from those of rrm3Delta cells. Rrm3p deletion derivatives that lacked smaller portions of the amino terminus were also defective, but the extent of replication pausing at tRNA genes, telomeres, and ribosomal DNA (rDNA) was not as great as in rrm3Delta cells. Deleting only 62 amino acids from the middle of the amino terminus affected only rDNA replication, suggesting that the amino terminus can confer locus-specific effects. Cells expressing a fusion protein consisting of the Rrm3p amino terminus and the Pif1p helicase domain displayed defects similar to rrm3Delta cells. These data demonstrate that the amino terminus of Rrm3p is essential for Rrm3p function. However, the helicase domain of Rrm3p also contributes to its functional specificity.
Figures





References
-
- Bennett, R. J., M. F. Noirot-Gros and J. C. Wang, 2000. Interaction between yeast sgs1 helicase and DNA topoisomerase III. J. Biol. Chem. 275: 26898–26905. - PubMed
-
- Bessler, J. B., J. Z. Torres and V. A. Zakian, 2001. The Pif1p subfamily of helicases: region specific DNA helicases. Trends Cell Biol. 11: 60–65. - PubMed
-
- Boeke, J. D., J. Trueheart, G. Natsoulis and G. R. Fink, 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154: 164–175. - PubMed
-
- Brewer, B. J., and W. L. Fangman, 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51: 463–471. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

