Drosophila klarsicht has distinct subcellular localization domains for nuclear envelope and microtubule localization in the eye
- PMID: 15579692
- PMCID: PMC1448802
- DOI: 10.1534/genetics.104.028662
Drosophila klarsicht has distinct subcellular localization domains for nuclear envelope and microtubule localization in the eye
Abstract
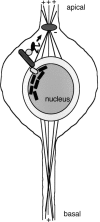
The Drosophila klarsicht (klar) gene is required for developmentally regulated migrations of photoreceptor cell nuclei in the eye. klar encodes a large ( approximately 250 kD) protein with only one recognizable amino acid sequence motif, a KASH (Klar, Anc-1, Syne-1 homology) domain, at its C terminus. It has been proposed that Klar facilitates nuclear migration by linking the nucleus to the microtubule organizing center (MTOC). Here we perform genetic and immunohistochemical experiments that provide a critical test of this model. We analyze mutants in the endogenous klar gene and also flies that express deleted forms of Klar protein from transgenes. We find that the KASH domain of Klar is critical for perinuclear localization and for function. In addition, we find that the N-terminal portion of Klar is also important for function and contains a domain that localizes the protein to microtubules apical to the nucleus. These results provide strong support for a model in which Klar links the nucleus to the MTOC.
Figures




Similar articles
-
Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye.Fly (Austin). 2007 Mar-Apr;1(2):75-85. doi: 10.4161/fly.4254. Fly (Austin). 2007. PMID: 18820457
-
Organelle-specific control of intracellular transport: distinctly targeted isoforms of the regulator Klar.Mol Biol Cell. 2005 Mar;16(3):1406-16. doi: 10.1091/mbc.e04-10-0920. Epub 2005 Jan 12. Mol Biol Cell. 2005. PMID: 15647372 Free PMC article.
-
The Drosophila KASH domain proteins Msp-300 and Klarsicht and the SUN domain protein Klaroid have no essential function during oogenesis.Fly (Austin). 2008 Mar-Apr;2(2):82-91. doi: 10.4161/fly.6288. Epub 2008 Mar 1. Fly (Austin). 2008. PMID: 18820478
-
Nuclear migration events throughout development.J Cell Sci. 2016 May 15;129(10):1951-61. doi: 10.1242/jcs.179788. J Cell Sci. 2016. PMID: 27182060 Free PMC article. Review.
-
LINC complex proteins in development and disease.Curr Top Dev Biol. 2014;109:287-321. doi: 10.1016/B978-0-12-397920-9.00004-4. Curr Top Dev Biol. 2014. PMID: 24947240 Review.
Cited by
-
Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules.J Cell Biol. 2012 Sep 3;198(5):833-46. doi: 10.1083/jcb.201204102. Epub 2012 Aug 27. J Cell Biol. 2012. PMID: 22927463 Free PMC article.
-
Another way to move chromosomes.Chromosoma. 2007 Dec;116(6):497-505. doi: 10.1007/s00412-007-0114-8. Epub 2007 Jul 18. Chromosoma. 2007. PMID: 17639451 Review.
-
Ca2+ channel-independent requirement for MAGUK family CACNB4 genes in initiation of zebrafish epiboly.Proc Natl Acad Sci U S A. 2008 Jan 8;105(1):198-203. doi: 10.1073/pnas.0707948105. Epub 2008 Jan 2. Proc Natl Acad Sci U S A. 2008. PMID: 18172207 Free PMC article.
-
Outer nuclear membrane protein Kuduk modulates the LINC complex and nuclear envelope architecture.J Cell Biol. 2017 Sep 4;216(9):2827-2841. doi: 10.1083/jcb.201606043. Epub 2017 Jul 17. J Cell Biol. 2017. PMID: 28716842 Free PMC article.
-
Making the LINC: SUN and KASH protein interactions.Biol Chem. 2015 Apr;396(4):295-310. doi: 10.1515/hsz-2014-0267. Biol Chem. 2015. PMID: 25720065 Free PMC article. Review.
References
-
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. - PubMed
-
- Apel, E. D., R. M. Lewis, R. M. Grady and J. R. Sanes, 2000. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J. Biol. Chem. 275: 31986–31995. - PubMed
-
- Fan, S.-S., and D. F. Ready, 1997. Glued participates in distinct microtubule-based activities in Drosophila eye development. Development 124: 1497–1507. - PubMed
-
- Fischer-Vize, J. A., and K. Mosley, 1994. marbles mutants: uncoupling cell determination and nuclear migration in the developing Drosophila eye. Development 120: 2609–2618. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

