Mutations in Su(var)205 and Su(var)3-7 suppress P-element-dependent silencing in Drosophila melanogaster
- PMID: 15579693
- PMCID: PMC1448784
- DOI: 10.1534/genetics.104.026914
Mutations in Su(var)205 and Su(var)3-7 suppress P-element-dependent silencing in Drosophila melanogaster
Abstract
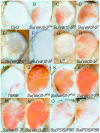
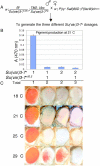
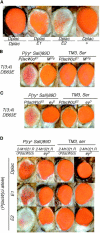
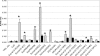
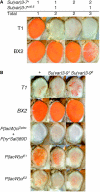
In Drosophila melanogaster, the w(+) transgene in P[lacW]ci(Dplac) is uniformly expressed throughout the adult eye. However, when other P elements are present, this w(+) transgene is randomly silenced and this produces a variegated eye phenotype. This P-element-dependent silencing (PDS) is limited to w(+) transgenes inserted in a specific region on chromosome 4. In a screen for genetic modifiers of PDS, we isolated mutations in Su(var)205, Su(var)3-7, and two unidentified genes that suppress this variegated phenotype. Therefore, only a few of the genes encoding heterochromatic modifiers act dose dependently in PDS. In addition, we recovered two spontaneous mutations of P[lacW]ci(Dplac) that variegate in the absence of P elements. These P[lacW]i(Dplac) derivatives have a gypsy element inserted proximally to the P[lacW]ci(Dplac) insert. The same mutations that suppress PDS also suppress w(+) silencing from these P[lacW]ci(Dplac) derivative alleles. This indicates that both cis-acting changes in sequence and trans-acting P elements cause a similar change in chromatin structure that silences w(+) expression in P[lacW]ci(Dplac). Together, these results confirm that PDS occurs at P[lacW]ci(Dplac) because of the chromatin structure at this chromosomal position. Studying w(+) variegation from P[lacW]ci(Dplac) provides a model for the interactions that can enhance heterochromatic silencing at single P-element inserts.
Figures





References
-
- Anxolabehere, D., M. G. Kidwell and G. Periquet, 1988. Molecular characteristics of diverse populations are consistent with the hypothesis of a recent invasion of Drosophila melanogaster by mobile P elements. Mol. Biol. Evol. 5: 252–269. - PubMed
-
- Ashburner, M., 1989a Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Ashburner, M., 1989b Drosophila: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Balasov, M. L., 2002. Genetic factors controlling white gene expression of the transposon A(R) 4-24 at a telomere in Drosophila melanogaster. Genome 45: 1025–1034. - PubMed
-
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas et al., 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

