Conserved function of medaka pink-eyed dilution in melanin synthesis and its divergent transcriptional regulation in gonads among vertebrates
- PMID: 15579703
- PMCID: PMC1448775
- DOI: 10.1534/genetics.104.030494
Conserved function of medaka pink-eyed dilution in melanin synthesis and its divergent transcriptional regulation in gonads among vertebrates
Abstract
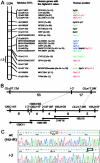
Medaka is emerging as a model organism for the study of vertebrate development and genetics, and its effectiveness in forward genetics should prove equal to that of zebrafish. Here, we identify by positional cloning a gene responsible for the medaka i-3 albino mutant. i-3 larvae have weakly tyrosinase-positive cells but lack strongly positive and dendritic cells, suggesting loss of fully differentiated melanophores. The region surrounding the i-3 locus is syntenic to human 19p13, but a BAC clone covering the i-3 locus contained orthologs located at 15q11-13, including OCA2 (P). Medaka P consists of 842 amino acids and shares approximately 65% identity with mammalian P proteins. The i-3 mutation is a four-base deletion in exon 13, which causes a frameshift and truncation of the protein. We detected medaka P transcripts in melanin-producing eyeballs and (putative) skin melanophores on embryos and an alternatively spliced form in the non-melanin-producing ovary or oocytes. The mouse p is similarly expressed in gonads, but not alternatively spliced. This is the first isolation of nonmammalian P, the functional mechanism of action of which has not yet been elucidated, even in mammals. Further investigation of the functions of P proteins and the regulation of their expression will provide new insight into body color determination and gene evolution.
Figures


Similar articles
-
The gsdf gene locus harbors evolutionary conserved and clustered genes preferentially expressed in fish previtellogenic oocytes.Gene. 2011 Feb 1;472(1-2):7-17. doi: 10.1016/j.gene.2010.10.014. Epub 2010 Oct 31. Gene. 2011. PMID: 21047546
-
Molecular cloning and gene expression of the prox1a and prox1b genes in the medaka, Oryzias latipes.Gene Expr Patterns. 2009 Jun;9(5):341-7. doi: 10.1016/j.gep.2009.02.004. Epub 2009 Feb 20. Gene Expr Patterns. 2009. PMID: 19233319
-
DMY is a Y-specific DM-domain gene required for male development in the medaka fish.Nature. 2002 May 30;417(6888):559-63. doi: 10.1038/nature751. Epub 2002 May 12. Nature. 2002. PMID: 12037570
-
Medaka genomics: a bridge between mutant phenotype and gene function.Mech Dev. 2004 Jul;121(7-8):619-28. doi: 10.1016/j.mod.2004.04.014. Mech Dev. 2004. PMID: 15210171 Review.
-
Regulation of germ cell sex identity in medaka.Curr Top Dev Biol. 2019;134:151-165. doi: 10.1016/bs.ctdb.2019.01.010. Epub 2019 Feb 18. Curr Top Dev Biol. 2019. PMID: 30999974 Review.
Cited by
-
Novel Functional Role of NK3R Expression in the Potentiating Effects on Somatolactin α Autoregulation in grass carp pituitary cells.Sci Rep. 2016 Oct 27;6:36102. doi: 10.1038/srep36102. Sci Rep. 2016. PMID: 27786296 Free PMC article.
-
OCA2 splice site variant in German Spitz dogs with oculocutaneous albinism.PLoS One. 2017 Oct 3;12(10):e0185944. doi: 10.1371/journal.pone.0185944. eCollection 2017. PLoS One. 2017. PMID: 28973042 Free PMC article.
-
Genomic Analysis of the Only Blind Cichlid Reveals Extensive Inactivation in Eye and Pigment Formation Genes.Genome Biol Evol. 2020 Aug 1;12(8):1392-1406. doi: 10.1093/gbe/evaa144. Genome Biol Evol. 2020. PMID: 32653909 Free PMC article.
-
The Genome of the Trinidadian Guppy, Poecilia reticulata, and Variation in the Guanapo Population.PLoS One. 2016 Dec 29;11(12):e0169087. doi: 10.1371/journal.pone.0169087. eCollection 2016. PLoS One. 2016. PMID: 28033408 Free PMC article.
-
ITRAQ Proteomic Analysis of Yellow and Black Skin in Jinbian Carp (Cyprinus carpio).Life (Basel). 2020 Sep 30;10(10):226. doi: 10.3390/life10100226. Life (Basel). 2020. PMID: 33007994 Free PMC article.
References
-
- Ancans, J., N. Flanagan, M. J. Hoogduijn and A. J. Thody, 2003. P-locus is a target for the melanogenic effects of MC-1R signaling: a possible control point for facultative pigmentation. Ann. NY Acad. Sci. 994: 373–377. - PubMed
-
- Fukamachi, S., A. Shimada and A. Shima, 2001. Mutations in the gene encoding B, a novel transporter protein, reduce melanin content in medaka. Nat. Genet. 28: 381–385. - PubMed
-
- Furutani-Seiki, M., T. Sasado, C. Morinaga, H. Suwa, K. Niwa et al., 2004. A systematic genome-wide screen for mutations affecting organogenesis in Medaka, Oryzias latipes. Mech. Dev. 121: 647–658. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

