The ubiquitin-proteasome pathway and plant development
- PMID: 15579807
- PMCID: PMC535867
- DOI: 10.1105/tpc.104.161220
The ubiquitin-proteasome pathway and plant development
Figures
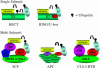

References
-
- Abel, S., Nguyen, M.D., and Theologis, A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251, 533–549. - PubMed
-
- Andrade, M.A., Gonzalez-Guzman, M., Serrano, R., and Rodriguez, P.L. (2001). A combination of the F-box motif and kelch repeats defines a large Arabidopsis family of F-box proteins. Plant Mol. Biol. 46, 603–614. - PubMed
-
- Azevedo, C., Santos-Rosa, M.J., and Shirasu, K. (2001). The U-box protein family in plants. Trends Plant Sci. 6, 354–358. - PubMed
-
- Bates, P.W., and Vierstra, R.D. (1999). UPL1 and 2, two 405 kDa ubiquitin-protein ligases from Arabidopsis thaliana related to the HECT-domain protein family. Plant J. 20, 183–195. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

