In silico study of amyloid beta-protein folding and oligomerization
- PMID: 15583128
- PMCID: PMC536046
- DOI: 10.1073/pnas.0408153101
In silico study of amyloid beta-protein folding and oligomerization
Abstract
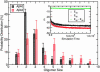
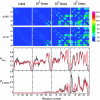
Experimental findings suggest that oligomeric forms of the amyloid beta protein (Abeta) play a critical role in Alzheimer's disease. Thus, elucidating their structure and the mechanisms of their formation is critical for developing therapeutic agents. We use discrete molecular dynamics simulations and a four-bead protein model to study oligomerization of two predominant alloforms, Abeta40 and Abeta42, at the atomic level. The four-bead model incorporates backbone hydrogen-bond interactions and amino acid-specific interactions mediated through hydrophobic and hydrophilic elements of the side chains. During the simulations we observe monomer folding and aggregation of monomers into oligomers of variable sizes. Abeta40 forms significantly more dimers than Abeta42, whereas pentamers are significantly more abundant in Abeta42 relative to Abeta40. Structure analysis reveals a turn centered at Gly-37-Gly-38 that is present in a folded Abeta42 monomer but not in a folded Abeta40 monomer and is associated with the first contacts that form during monomer folding. Our results suggest that this turn plays an important role in Abeta42 pentamer formation. Abeta pentamers have a globular structure comprising hydrophobic residues within the pentamer's core and hydrophilic N-terminal residues at the surface of the pentamer. The N termini of Abeta40 pentamers are more spatially restricted than Abeta42 pentamers. Abeta40 pentamers form a beta-strand structure involving Ala-2-Phe-4, which is absent in Abeta42 pentamers. These structural differences imply a different degree of hydrophobic core exposure between pentamers of the two alloforms, with the hydrophobic core of the Abeta42 pentamer being more exposed and thus more prone to form larger oligomers.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

