Sequencing needs for viral diagnostics
- PMID: 15583268
- PMCID: PMC535215
- DOI: 10.1128/JCM.42.12.5472-5476.2004
Sequencing needs for viral diagnostics
Abstract
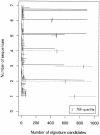
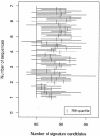
We built a system to guide decisions regarding the amount of genomic sequencing required to develop diagnostic DNA signatures, which are short sequences that are sufficient to uniquely identify a viral species. We used our existing DNA diagnostic signature prediction pipeline, which selects regions of a target species genome that are conserved among strains of the target (for reliability, to prevent false negatives) and unique relative to other species (for specificity, to avoid false positives). We performed simulations, based on existing sequence data, to assess the number of genome sequences of a target species and of close phylogenetic relatives (near neighbors) that are required to predict diagnostic signature regions that are conserved among strains of the target species and unique relative to other bacterial and viral species. For DNA viruses such as variola (smallpox), three target genomes provide sufficient guidance for selecting species-wide signatures. Three near-neighbor genomes are critical for species specificity. In contrast, most RNA viruses require four target genomes and no near-neighbor genomes, since lack of conservation among strains is more limiting than uniqueness. Severe acute respiratory syndrome and Ebola Zaire are exceptional, as additional target genomes currently do not improve predictions, but near-neighbor sequences are urgently needed. Our results also indicate that double-stranded DNA viruses are more conserved among strains than are RNA viruses, since in most cases there was at least one conserved signature candidate for the DNA viruses and zero conserved signature candidates for the RNA viruses.
Figures

References
-
- Fitch, J. P., B. A. Chromy, C. E. Forde, E. Garcia, S. N. Gardner, P. Gu, T. A. Kuczmarksi, C. Melius, S. L. McCutchen-Maloney, F. M. Milanovich, V. L. Motin, L. L. Ott, A. Quong, J. Quong, J. M. Rocco, T. R. Slezak, B. A. Sokhansanj, E. A. Vitalis, A. T. Zemla, and P. M. McCready. 2002. Presented at the IEEE Workshop on Genomic Signal Processing and Statistics (GENSIPS), Oct. 12-13, 2002, Raleigh, N.C.
-
- Fitch, J. P., S. N. Gardner, T. A. Kuczmarski, S. Kurtz, R. Myers, L. L. Ott, T. R. Slezak, E. A. Vitalis, A. T. Zemla, and P. M. McCready. 2002. Rapid development of nucleic acid diagnostics. Proc. IEEE 90:1708-1721.
-
- Giegerich, R., S. Kurtz, and J. Stoye. 2003. Efficient implementation of lazy suffix trees. Softw. Pract. Exp. 33:1035-1049.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

