Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment
- PMID: 15583313
- PMCID: PMC535267
- DOI: 10.1128/JCM.42.12.5783-5792.2004
Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment
Abstract
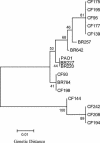
Pseudomonas aeruginosa is a gram-negative rod that is ubiquitous in nature. P. aeruginosa is also the quintessential opportunistic pathogen, causing a wide variety of infections in compromised hosts. In cystic fibrosis patients, P. aeruginosa is the leading cause of death. In this study, the evolutionary genetic relationships among 17 P. aeruginosa isolates were examined by comparative sequence analysis of the housekeeping gene encoding malate dehydrogenase and the chaperone groEL. The P. aeruginosa isolates examined included the sequenced strain PAO1, 11 strains recovered from cystic fibrosis patients in Ireland, 4 environmental isolates recovered from a hospital environment, and 1 isolate recovered from a plant rhizosphere. Phylogenetically, clinical and environmental isolates clustered together with one another on the mdh gene tree. At the groEL locus, among the 17 isolates examined, only two polymorphic sites were observed, highlighting the close genetic relationship between isolates from these different environments. Phenotypic analysis of 12 traits among our isolates, however, found that only clinical isolates produced phenazines and elastase. Furthermore, molecular analysis of the distribution of 15 regions associated with virulence showed that two of the environmental isolates examined lacked the majority of regions. Among the clinical isolates examined, the 15 virulence regions were variably present. The distribution of two prophages (Bacto1, Pf1) was also determined, with most isolates encoding both these regions. Of the four genomic islands (the flagellum island and PAGI-1, -2, and -3) examined, only two isolates contained the flagellum island, and PAGI-1, -2, and -3 were absent from all isolates tested. Our data demonstrate the significant role horizontal gene transfer and recombination, together with gene loss, play in the evolution of this important human pathogen.
Figures


Similar articles
-
A cystic fibrosis epidemic strain of Pseudomonas aeruginosa displays enhanced virulence and antimicrobial resistance.J Bacteriol. 2005 Jul;187(14):4908-20. doi: 10.1128/JB.187.14.4908-4920.2005. J Bacteriol. 2005. PMID: 15995206 Free PMC article.
-
Comparison of genome fingerprinting with conventional typing methods used on Pseudomonas aeruginosa isolates from cystic fibrosis patients.APMIS. 1993 Feb;101(2):168-75. APMIS. 1993. PMID: 8098219
-
Use of phage display to identify potential Pseudomonas aeruginosa gene products relevant to early cystic fibrosis airway infections.Infect Immun. 2005 Jan;73(1):444-52. doi: 10.1128/IAI.73.1.444-452.2005. Infect Immun. 2005. PMID: 15618183 Free PMC article.
-
Polyagglutinable Pseudomonas aeruginosa from cystic fibrosis patients. A survey.APMIS Suppl. 1994;46:1-44. APMIS Suppl. 1994. PMID: 7811529 Review.
-
[Various typing methods of Pseudomonas aeruginosa strains isolated from cystic fibrosis patients].Med Mal Infect. 2008 May;38(5):238-47. doi: 10.1016/j.medmal.2008.02.005. Epub 2008 Apr 3. Med Mal Infect. 2008. PMID: 18394837 Review. French.
Cited by
-
A novel phenolic derivative inhibits AHL-dependent quorum sensing signaling in Pseudomonas aeruginosa.Front Pharmacol. 2022 Sep 20;13:996871. doi: 10.3389/fphar.2022.996871. eCollection 2022. Front Pharmacol. 2022. PMID: 36204236 Free PMC article.
-
Filamentous Bacteriophage Produced by Pseudomonas aeruginosa Alters the Inflammatory Response and Promotes Noninvasive Infection In Vivo.Infect Immun. 2016 Dec 29;85(1):e00648-16. doi: 10.1128/IAI.00648-16. Print 2017 Jan. Infect Immun. 2016. PMID: 27795361 Free PMC article.
-
The Effect of Silver Nanoparticles on Pyocyanin Production of Pseudomonas aeruginosa Isolated From Clinical Specimens.Avicenna J Med Biotechnol. 2021 Apr-Jun;13(2):98-103. doi: 10.18502/ajmb.v13i2.5529. Avicenna J Med Biotechnol. 2021. PMID: 34012526 Free PMC article.
-
Filamentous prophage Pf4 promotes genetic exchange in Pseudomonas aeruginosa.ISME J. 2024 Jan 8;18(1):wrad025. doi: 10.1093/ismejo/wrad025. ISME J. 2024. PMID: 38365255 Free PMC article.
-
High frequency of the exoU+/exoS+ genotype associated with multidrug-resistant "high-risk clones" of Pseudomonas aeruginosa clinical isolates from Peruvian hospitals.Sci Rep. 2019 Jul 26;9(1):10874. doi: 10.1038/s41598-019-47303-4. Sci Rep. 2019. PMID: 31350412 Free PMC article.
References
-
- Adams, C., M. Morris-Quinn, F. McConnell, J. West, B. Lucey, C. Shortt, B. Cryan, J. B. Watson, and F. O'Gara. 1998. Epidemiology and clinical impact of Pseudomonas aeruginosa infection in cystic fibrosis using AP-PCR fingerprinting. J. Infect. 37:151-158. - PubMed
-
- Boyd, E. F., and H. Brussow. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521-529. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

