Mapping of genomic segments of influenza B virus strains by an oligonucleotide microarray method
- PMID: 15583314
- PMCID: PMC535258
- DOI: 10.1128/JCM.42.12.5793-5801.2004
Mapping of genomic segments of influenza B virus strains by an oligonucleotide microarray method
Abstract
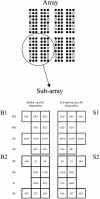
Similar to other segmented RNA viruses, influenza viruses can exchange genome segments and form a wide variety of reassortant strains upon coreplication within a host cell. Therefore, the mapping of genome segments of influenza viruses is essential for understanding their phenotypes. In this work, we have developed an oligonucleotide microarray hybridization method for simultaneous genotyping of all genomic segments of two highly homologous strains of influenza B virus. A few strain-specific oligonucleotide probes matching each of the eight segments of the viral genomes of the B/Beijing/184/93 and B/Shangdong/7/97 strains were hybridized with PCR-amplified fluorescently labeled single-stranded DNA. Even though there were a few mismatches among the genomes of the studied virus strains, microarray hybridization showed highly significant and reproducible discrimination ability and allowed us to determine the origins of individual genomic segments in a series of reassortant strains prepared as vaccine candidates. Additionally, we were able to detect the presence of at least 5% of mixed genotypes in virus stocks even when conventional sequencing methods failed, for example, for the NS segment. Thus, the proposed microarray method can be used for (i) rapid and reliable genome mapping of highly homologous influenza B viruses and (ii) extensive monitoring of influenza B virus reassortants and the mixed genotypes. The array can be expanded by adding new oligoprobes and using more quantitative assays to determine the origin of individual genomic segments in series of reassortant strains prepared as vaccine candidates or in mixed virus populations.
Figures



References
-
- Cha, T. A., J. Zhao, E. Lane, M. A. Murray, and D. S. Stec. 1997. Determination of the genome composition of influenza virus reassortants using multiplex reverse transcription-polymerase chain reaction followed by fluorescent single-strand conformation polymorphism analysis. Anal. Biochem. 252:24-32. - PubMed
-
- Chanock, R. M., and B. R. Murphy. 1980. Use of temperature-sensitive and cold-adapted mutant viruses in immunoprophylaxis of acute respiratory tract disease. Rev. Infect. Dis. 2:421-432. - PubMed
-
- Chare, E. R., E. A. Gould, and E. C. Holmes. 2003. Phylogenetic analysis reveals a low rate of homologous recombination in negative-sense RNA viruses. J. Gen. Virol. 84:2691-2703. - PubMed
-
- Chen, H., Y. Matsuoka, D. Swayne, Q. Chen, N. J. Cox, B. R. Murphy, and K. Subbarao. 2003. Generation and characterization of a cold-adapted influenza A H9N2 reassortant as a live pandemic influenza virus vaccine candidate. Vaccine 21:4430-4436. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

