Algorithmic self-assembly of DNA Sierpinski triangles
- PMID: 15583715
- PMCID: PMC534809
- DOI: 10.1371/journal.pbio.0020424
Algorithmic self-assembly of DNA Sierpinski triangles
Abstract
Algorithms and information, fundamental to technological and biological organization, are also an essential aspect of many elementary physical phenomena, such as molecular self-assembly. Here we report the molecular realization, using two-dimensional self-assembly of DNA tiles, of a cellular automaton whose update rule computes the binary function XOR and thus fabricates a fractal pattern--a Sierpinski triangle--as it grows. To achieve this, abstract tiles were translated into DNA tiles based on double-crossover motifs. Serving as input for the computation, long single-stranded DNA molecules were used to nucleate growth of tiles into algorithmic crystals. For both of two independent molecular realizations, atomic force microscopy revealed recognizable Sierpinski triangles containing 100-200 correct tiles. Error rates during assembly appear to range from 1% to 10%. Although imperfect, the growth of Sierpinski triangles demonstrates all the necessary mechanisms for the molecular implementation of arbitrary cellular automata. This shows that engineered DNA self-assembly can be treated as a Turing-universal biomolecular system, capable of implementing any desired algorithm for computation or construction tasks.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures
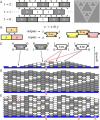





Comment in
-
The emergence of complexity: lessons from DNA.PLoS Biol. 2004 Dec;2(12):e431. doi: 10.1371/journal.pbio.0020431. PLoS Biol. 2004. PMID: 15597116 Free PMC article.
References
-
- Adleman LM. Molecular computation of solutions to combinatorial problems. Science. 1994;266:1021–1024. - PubMed
-
- Adleman LM, Cheng Q, Goel A, Huang MDA. Running time and program size for self-assembled squares. Symposium on Theory of Computing (STOC) New York: Association for Computing Machinery; 2001. pp. 740–748.
-
- Adleman L, Kari J, Kari L, Reishus D. On the decidability of self-assembly of infinite ribbons. Symposium on Foundations of Computer Science (FOCS) New York: Association for Computing Machinery; 2002. pp. 530–537.
-
- Bennett CH. Logical depth and physical complexity. In: Herken R, editor. The universal Turing machine: A half-century survey. Vienna: Springer-Verlag; 1995. pp. 207–235.
-
- Bloomfield VA, Crothers DM, Tinoco I. Nucleic acids: Structures, properties, and functions. Sausalito, California: University Science Books; 2000. 672 pp.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

