Identification of small-molecule antagonists that inhibit an activator: coactivator interaction
- PMID: 15585582
- PMCID: PMC539725
- DOI: 10.1073/pnas.0406374101
Identification of small-molecule antagonists that inhibit an activator: coactivator interaction
Abstract
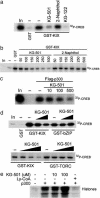
Phosphorylation of the cAMP response element binding protein (CREB) at Ser-133 in response to hormonal stimuli triggers cellular gene expression via the recruitment of the histone acetylase coactivator paralogs CREB binding protein (CBP) and p300 to the promoter. The NMR structure of the CREB:CBP complex, using relevant interaction domains called KID and KIX, respectively, reveals a shallow hydrophobic groove on the surface of KIX that accommodates an amphipathic helix in phospho (Ser-133) KID. Using an NMR-based screening approach on a preselected small-molecule library, we identified several compounds that bind to different surfaces on KIX. One of these, KG-501 (2-naphthol-AS-E-phosphate), targeted a surface distal to the CREB binding groove that includes Arg-600, a residue that is required for the CREB:CBP interaction. When added to live cells, KG-501 disrupted the CREB: CBP complex and attenuated target gene induction in response to cAMP agonist. These results demonstrate the ability of small molecules to interfere with second-messenger signaling cascades by inhibiting specific protein-protein interactions in the nucleus.
Figures


References
-
- Ptashne, M. & Gann, A. (1997) Nature 386, 569–577. - PubMed
-
- Radhakrishnan, I., Perez-Alvarado, G. C., Parker, D., Dyson, H. J., Montminy, M. & Wright, P. E. (1997) Cell 91, 741–752. - PubMed
-
- Radhakrishnan, I., Perez-Alvarado, G. C., Parker, D., Dyson, H. J., Montminy, M. R. & Wright, P. E. (1999) J. Mol. Biol. 287, 859–865. - PubMed
-
- Parker, D., Jhala, U., Radhakrishnan, I., Yaffe, M., Reyes, C., Shulman, A., Cantley, L., Wright, P. & Montminy, M. (1998) Mol. Cell 2, 353–359. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

