Optimized LOWESS normalization parameter selection for DNA microarray data
- PMID: 15588297
- PMCID: PMC539276
- DOI: 10.1186/1471-2105-5-194
Optimized LOWESS normalization parameter selection for DNA microarray data
Abstract
Background: Microarray data normalization is an important step for obtaining data that are reliable and usable for subsequent analysis. One of the most commonly utilized normalization techniques is the locally weighted scatterplot smoothing (LOWESS) algorithm. However, a much overlooked concern with the LOWESS normalization strategy deals with choosing the appropriate parameters. Parameters are usually chosen arbitrarily, which may reduce the efficiency of the normalization and result in non-optimally normalized data. Thus, there is a need to explore LOWESS parameter selection in greater detail.
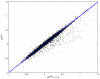
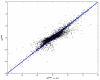
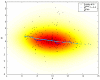
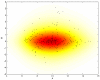
Results and discussion: In this work, we discuss how to choose parameters for the LOWESS method. Moreover, we present an optimization approach for obtaining the fraction of data points utilized in the local regression and analyze results for local print-tip normalization. The optimization procedure determines the bandwidth parameter for the local regression by minimizing a cost function that represents the mean-squared difference between the LOWESS estimates and the normalization reference level. We demonstrate the utility of the systematic parameter selection using two publicly available data sets. The first data set consists of three self versus self hybridizations, which allow for a quantitative study of the optimization method. The second data set contains a collection of DNA microarray data from a breast cancer study utilizing four breast cancer cell lines. Our results show that different parameter choices for the bandwidth window yield dramatically different calibration results in both studies.
Conclusions: Results derived from the self versus self experiment indicate that the proposed optimization approach is a plausible solution for estimating the LOWESS parameters, while results from the breast cancer experiment show that the optimization procedure is readily applicable to real-life microarray data normalization. In summary, the systematic approach to obtain critical parameters in the LOWESS technique is likely to produce data that optimally meets assumptions made in the data preprocessing step and thereby makes studies utilizing the LOWESS method unambiguous and easier to repeat.
Figures

References
-
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. - PubMed

