The functional landscape of mouse gene expression
- PMID: 15588312
- PMCID: PMC549719
- DOI: 10.1186/jbiol16
The functional landscape of mouse gene expression
Abstract
Background: Large-scale quantitative analysis of transcriptional co-expression has been used to dissect regulatory networks and to predict the functions of new genes discovered by genome sequencing in model organisms such as yeast. Although the idea that tissue-specific expression is indicative of gene function in mammals is widely accepted, it has not been objectively tested nor compared with the related but distinct strategy of correlating gene co-expression as a means to predict gene function.
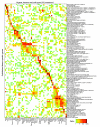
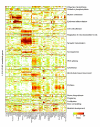
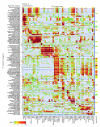
Results: We generated microarray expression data for nearly 40,000 known and predicted mRNAs in 55 mouse tissues, using custom-built oligonucleotide arrays. We show that quantitative transcriptional co-expression is a powerful predictor of gene function. Hundreds of functional categories, as defined by Gene Ontology 'Biological Processes', are associated with characteristic expression patterns across all tissues, including categories that bear no overt relationship to the tissue of origin. In contrast, simple tissue-specific restriction of expression is a poor predictor of which genes are in which functional categories. As an example, the highly conserved mouse gene PWP1 is widely expressed across different tissues but is co-expressed with many RNA-processing genes; we show that the uncharacterized yeast homolog of PWP1 is required for rRNA biogenesis.
Conclusions: We conclude that 'functional genomics' strategies based on quantitative transcriptional co-expression will be as fruitful in mammals as they have been in simpler organisms, and that transcriptional control of mammalian physiology is more modular than is generally appreciated. Our data and analyses provide a public resource for mammalian functional genomics.
Figures





Comment in
-
All systems GO for understanding mouse gene function.J Biol. 2004;3(5):20. doi: 10.1186/jbiol19. Epub 2004 Dec 17. J Biol. 2004. PMID: 15610553 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

