The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs
- PMID: 15588487
- PMCID: PMC3525267
- DOI: 10.1186/1479-7364-1-4-274
The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs
Abstract
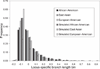
Understanding the nature of evolutionary relationships among persons and populations is important for the efficient application of genome science to biomedical research. We have analysed 8,525 autosomal single nucleotide polymorphisms (SNPs) in 84 individuals from four populations: African-American, European-American, Chinese and Japanese. Individual relationships were reconstructed using the allele sharing distance and the neighbour-joining tree making method. Trees show clear clustering according to population, with the root branching from the African-American clade. The African-American cluster is much less star-like than European-American and East Asian clusters, primarily because of admixture. Furthermore, on the East Asian branch, all ten Chinese individuals cluster together and all ten Japanese individuals cluster together. Using positional information, we demonstrate strong correlations between inter-marker distance and both locus-specific FST (the proportion of total variation due to differentiation) levels and branch lengths. Chromosomal maps of the distribution of locus-specific branch lengths were constructed by combining these data with other published SNP markers (total of 33,704 SNPs). These maps clearly illustrate a non-uniform distribution of human genetic substructure, an instructional and useful paradigm for education and research.
Figures
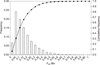
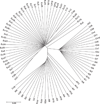
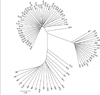




References
-
- Schork N, Fallin D, Tiwari HK, In: Handbook of Statistical Genetics. Balding, D., Bishop, M. and Cannings, C, editor. John Wiley and Sons, Hoboken, NJ; 2001. 'Pharmacogenetics'; pp. 741–764.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

