Relaxed primer specificity associated with reverse transcriptases encoded by the pFOXC retroplasmids of Fusarium oxysporum
- PMID: 15590832
- PMCID: PMC539014
- DOI: 10.1128/EC.3.6.1589-1600.2004
Relaxed primer specificity associated with reverse transcriptases encoded by the pFOXC retroplasmids of Fusarium oxysporum
Abstract
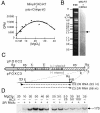
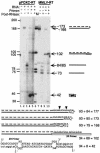
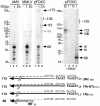
The pFOXC mitochondrial retroplasmids are small, autonomously replicating linear DNAs that have a telomere-like repeat of a 5-bp sequence at their termini. The plasmids are possible evolutionary precursors of the ribonucleoprotein complex telomerase, as they encode an active reverse transcriptase (RT) that is involved in plasmid replication. Using an in vitro system to study reverse transcription, we show that the pFOXC RT is capable of copying in vitro-synthesized RNAs by use of cDNA primers or extension of snapped-back RNA templates. The ability of the pFOXC RT to use base-paired primers distinguishes it from the closely related RTs encoded by the Mauriceville and Varkud mitochondrial retroplasmids of Neurospora spp. Reaction products are similar, but not identical, to those obtained with conventional RTs, and differences reflect the ability of the pFOXC RT to initiate cDNA synthesis with loosely associated primers. The pFOXC RT can also copy DNA templates and extend 3' mismatched DNA oligonucleotide primers. Analysis of pFOXC in vivo replication intermediates suggests that telomeric repeats are added during reverse transcription, and the ability to extend loosely associated primers could play a role in repeat formation by mechanisms similar to those associated with telomerase and certain non-long-terminal-repeat retrotransposons.
Figures



References
-
- Akins, R. A., D. M. Grant, L. L. Stohl, D. A. Bottorff, F. E. Nargang, and A. M. Lambowitz. 1988. Nucleotide sequence of the Varkud mitochondrial plasmid of Neurospora and synthesis of a 5′ transcript with a 5′ leader derived from mitochondrial RNA. J. Mol. Biol. 204:1-25. - PubMed
-
- Antal, Z., L. Manczinger, L. Kredics, F. Kevei, and E. Nagy. 2002. Complete DNA sequence and analysis of a mitochondrial plasmid in the mycoparasitic Trichoderma harzianum strain T95. Plasmid 47:148-152. - PubMed
-
- Chen, B., and A. M. Lambowitz. 1997. De novo and DNA primer-mediated initiation of cDNA synthesis by the Mauriceville retroplasmid reverse transcriptase involve recognition of a 3′ CCA sequence. J. Mol. Biol. 271:311-332. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

