The small-subunit processome is a ribosome assembly intermediate
- PMID: 15590835
- PMCID: PMC539036
- DOI: 10.1128/EC.3.6.1619-1626.2004
The small-subunit processome is a ribosome assembly intermediate
Abstract
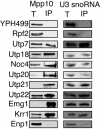
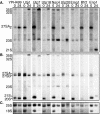
The small-subunit (SSU) processome is a large ribonucleoprotein required for the biogenesis of the 18S rRNA and likely corresponds to the terminal knobs visualized by electron microscopy on the 5' end of nascent rRNAs. The original purification of the SSU processome of Saccharomyces cerevisiae resulted in the identification of 28 proteins. Here, we characterize 12 additional protein components, including five small-ribosomal-subunit proteins (Rps4, Rps6, Rps7, Rps9, and Rps14) that had previously been copurified. Our multiple criteria for including a component as a bona fide SSU processome component included coimmunoprecipitation with Mpp10 (an SSU processome component), the U3 snoRNA, and the anticipated pre-rRNAs. Importantly, the association of specific ribosomal proteins with the SSU processome suggests that the SSU processome has roles in both pre-rRNA processing and ribosome assembly. These ribosomal proteins may be analogous to the primary or secondary RNA binding proteins first described in bacterial in vitro ribosome assembly maps. In addition to the ribosomal proteins and based on the same experimental approach, we found seven other proteins (Utp18, Noc4, Utp20, Utp21, Utp22, Emg1, and Krr1) to be bona fide SSU processome proteins.
Figures




References
-
- Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

