A novel E2 box-GATA element modulates Cdc6 transcription during human cells polyploidization
- PMID: 15590906
- PMCID: PMC535689
- DOI: 10.1093/nar/gkh981
A novel E2 box-GATA element modulates Cdc6 transcription during human cells polyploidization
Abstract
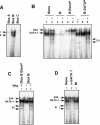
Cdc6 is a key regulator of the strict alternation of S and M phases during the mitotic cell cycle. In mammalian and plant cells that physiologically become polyploid, cdc6 is transcriptionally and post-translationally regulated. We have recently reported that Cdc6 levels are maintained in megakaryoblastic HEL cells, but severely downregulated by ectopic expression of transcriptional repressor Drosophila melanogaster escargot. Here, we show that cdc6 promoter activity is upregulated during megakaryocytic differentiation of HEL endoreplicating cells, and that Escargot interferes with such activation. Transactivation experiments showed that a 1.7 kb region located at 2800 upstream cdc6 transcription initiation site behaved as a potent enhancer in endoreplicating cells only. This activity was mainly dependent on a novel cis-regulatory element composed by an E2 box overlapping a GATA motif. Ectopic Escargot could bind this regulatory element in vitro and endogenous GATA-1 and E2A formed specific complexes in megakaryoblastic cells as well as in primary megakaryocytes. Chromatin Immunoprecipitation analysis revealed that both transcription factors were occupying the E2 box/GATA site in vivo. Altogether, these data suggest that cdc6 expression could be actively maintained during megakaryocytic differentiation through transcriptional mechanisms involving specific cis- and trans-regulatory elements.
Figures



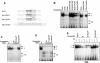



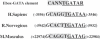
Similar articles
-
CDC6 expression is regulated by lineage-specific transcription factor GATA1.Cell Cycle. 2012 Aug 15;11(16):3055-66. doi: 10.4161/cc.21471. Epub 2012 Aug 8. Cell Cycle. 2012. PMID: 22871742 Free PMC article.
-
Regulation of CDC6, geminin, and CDT1 in human cells that undergo polyploidization.Mol Biol Cell. 2002 Nov;13(11):3989-4000. doi: 10.1091/mbc.e02-04-0217. Mol Biol Cell. 2002. PMID: 12429841 Free PMC article.
-
Heterologous expression of the transcriptional regulator escargot inhibits megakaryocytic endomitosis.J Biol Chem. 2001 Nov 16;276(46):43413-8. doi: 10.1074/jbc.M106006200. Epub 2001 Aug 9. J Biol Chem. 2001. PMID: 11498537
-
The Hematopoietic Stem and Progenitor Cell Cistrome: GATA Factor-Dependent cis-Regulatory Mechanisms.Curr Top Dev Biol. 2016;118:45-76. doi: 10.1016/bs.ctdb.2016.01.002. Epub 2016 Feb 26. Curr Top Dev Biol. 2016. PMID: 27137654 Free PMC article. Review.
-
Regulation of the transcription factor GATA-1 at the gene and protein level.Cell Mol Life Sci. 2001 Dec;58(14):2008-17. doi: 10.1007/PL00000833. Cell Mol Life Sci. 2001. PMID: 11814053 Free PMC article. Review.
Cited by
-
Endoreplication: polyploidy with purpose.Genes Dev. 2009 Nov 1;23(21):2461-77. doi: 10.1101/gad.1829209. Genes Dev. 2009. PMID: 19884253 Free PMC article. Review.
-
E2A Modulates Stemness, Metastasis, and Therapeutic Resistance of Breast Cancer.Cancer Res. 2021 Sep 1;81(17):4529-4544. doi: 10.1158/0008-5472.CAN-20-2685. Epub 2021 Jun 18. Cancer Res. 2021. PMID: 34145034 Free PMC article.
-
Enhancement of PRMT6 binding to a novel germline GATA1 mutation associated with congenital anemia.Haematologica. 2024 Sep 1;109(9):2955-2968. doi: 10.3324/haematol.2023.284183. Haematologica. 2024. PMID: 38385251 Free PMC article.
-
CDC6 expression is regulated by lineage-specific transcription factor GATA1.Cell Cycle. 2012 Aug 15;11(16):3055-66. doi: 10.4161/cc.21471. Epub 2012 Aug 8. Cell Cycle. 2012. PMID: 22871742 Free PMC article.
-
The SCL +40 enhancer targets the midbrain together with primitive and definitive hematopoiesis and is regulated by SCL and GATA proteins.Mol Cell Biol. 2007 Oct;27(20):7206-19. doi: 10.1128/MCB.00931-07. Epub 2007 Aug 20. Mol Cell Biol. 2007. PMID: 17709394 Free PMC article.
References
-
- Edgar B.A. and Orr-Weaver,T.L. (2001) Endoreplication cell cycles: more for less, Cell, 105, 297–306. - PubMed
-
- Ravid K., Lu,J., Zimmet,J.M. and Jones,M.R. (2002) Roads to polyploidy: the megakaryocyte example. J. Cell Physiol., 190, 7–20. - PubMed
-
- Zimmet J. and Ravid,K. (2000) Polyploidy: occurrence in nature, mechanisms, and significance for the megakaryocyte-platelet system. Exp. Hematol., 1, 3–16. - PubMed
-
- Garcia P. and Cales,C. (1996) Endoreplication in megakaryoblastic cell lines is accompanied by sustained expression of G1/S cyclins and downregulation of cdc25C. Oncogene, 13, 695–703. - PubMed

