Structural and functional analysis of a 0.5-Mb chicken region orthologous to the imprinted mammalian Ascl2/Mash2-Igf2-H19 region
- PMID: 15590938
- PMCID: PMC540284
- DOI: 10.1101/gr.2609605
Structural and functional analysis of a 0.5-Mb chicken region orthologous to the imprinted mammalian Ascl2/Mash2-Igf2-H19 region
Abstract
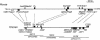
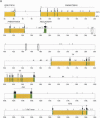
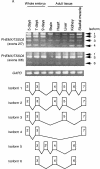
Previous studies revealed that Igf2 and Mpr/Igf2r are imprinted in eutherian mammals and marsupials but not in monotremes or birds. Igf2 lies in a large imprinted cluster in eutherians, and its imprinting is regulated by long-range mechanisms. As a step to understand how the imprinted cluster evolved, we have determined a 490-kb chicken sequence containing the orthologs of mammalian Ascl2/Mash2, Ins2 and Igf2. We found that most of the genes in this region are conserved between chickens and mammals, maintaining the same transcriptional polarities and exon-intron structures. However, H19, an imprinted noncoding transcript, was absent from the chicken sequence. Chicken ASCL2/CASH4 and INS, the orthologs of the imprinted mammalian genes, showed biallelic expression, further supporting the notion that imprinting evolved after the divergence of mammals and birds. The H19 imprinting center and many of the local regulatory elements identified in mammals were not found in chickens. Also, a large segment of tandem repeats and retroelements identified between the two imprinted subdomains in mice was not found in chickens. Our findings show that the imprinted genes were clustered before the emergence of imprinting and that the elements associated with imprinting probably evolved after the divergence of mammals and birds.
Figures
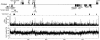





References
-
- Ainscough, J.F-X., John, R.M., Barton, S.C., and Surani, M.A. 2000. A skeletal muscle-specific mouse Igf2 repressor lies 40 kb downstream of the gene. Development 127: 3923-3930. - PubMed
-
- Andria, M.L., Hsieh, C.L., Oren, R., Francke, U., and Levy, S. 1991. Genomic organization and chromosomal localization of the TAPA-1 gene. J. Immunol. 147: 1030-1036. - PubMed
-
- Barlow, D.P. 1993. Methylation and imprinting: From host defense to gene regulation. Science 260: 309-310. - PubMed
-
- Bell, A.C. and Felsenfeld, G. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482-485. - PubMed
Web site references
-
- http://c3.biomath.mssm.edu/trf.html; Tandem Repeats Finder.
-
- http://genes.mit.edu/GENSCAN.html; GENSCAN.
-
- http://nog.cse.psu.edu/pipmaker/; PipMaker.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
