Evolution and functional classification of vertebrate gene deserts
- PMID: 15590943
- PMCID: PMC540279
- DOI: 10.1101/gr.3015505
Evolution and functional classification of vertebrate gene deserts
Abstract
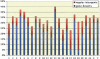
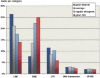
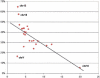
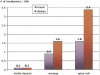
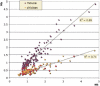
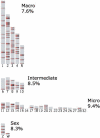
Large tracts of the human genome, known as gene deserts, are devoid of protein-coding genes. Dichotomy in their level of conservation with chicken separates these regions into two distinct categories, stable and variable. The separation is not caused by differences in rates of neutral evolution but instead appears to be related to different biological functions of stable and variable gene deserts in the human genome. Gene Ontology categories of the adjacent genes are strongly biased toward transcriptional regulation and development for the stable gene deserts, and toward distinctively different functions for the variable gene deserts. Stable gene deserts resist chromosomal rearrangements and appear to harbor multiple distant regulatory elements physically linked to their neighboring genes, with the linearity of conservation invariant throughout vertebrate evolution.
Figures
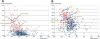
References
-
- Bell, A.C., West, A.G., and Felsenfeld, G. 2001. Insulators and boundaries: Versatile regulatory elements in the eukaryotic. Science 291: 447-450. - PubMed
-
- Carter, D., Chakalova, L., Osborne, C.S., Dai, Y.F., and Fraser, P. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32: 623-626. - PubMed
-
- Dehal, P., Predki, P., Olsen, A.S., Kobayashi, A., Folta, P., Lucas, S., Land, M., Terry, A., Ecale Zhou, C.L., Rash, S., et al. 2001. Human chromosome 19 and related regions in mouse: Conservative and lineage-specific evolution. Science 293: 104-111. - PubMed
-
- Dorsett, D. 1999. Distant liaisons: Long-range enhancer-promoter interactions in Drosophila. Curr. Opin. Genet. Dev. 9: 505-514. - PubMed
Web site references
-
- http://ecrbrowser.dcode.org; ECR Browser.
-
- http://genome.ucsc.edu; UCSC Genome database.
-
- http://hgdownload.cse.ucsc.edu/goldenPath/hg16/regPotential/; data used to generate the regulatory potential tracks.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
