Comparison of the chicken and turkey genomes reveals a higher rate of nucleotide divergence on microchromosomes than macrochromosomes
- PMID: 15590944
- PMCID: PMC540272
- DOI: 10.1101/gr.3021305
Comparison of the chicken and turkey genomes reveals a higher rate of nucleotide divergence on microchromosomes than macrochromosomes
Abstract
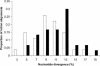
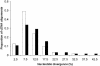
A distinctive feature of the avian genome is the large heterogeneity in the size of chromosomes, which are usually classified into a small number of macrochromosomes and numerous microchromosomes. These chromosome classes show characteristic differences in a number of interrelated features that could potentially affect the rate of sequence evolution, such as GC content, gene density, and recombination rate. We studied the effects of these factors by analyzing patterns of nucleotide substitution in two sets of chicken-turkey sequence alignments. First, in a set of 67 orthologous introns, divergence was significantly higher in microchromosomes (chromosomes 11-38; 11.7% divergence) than in both macrochromosomes (chromosomes 1-5; 9.9% divergence; P = 0.016) and intermediate-sized chromosomes (chromosomes 6-10; 9.5% divergence; P = 0.026). At least part of this difference was due to the higher incidence of CpG sites on microchromosomes. Second, using 155 orthologous coding sequences we noted a similar pattern, in which synonymous substitution rates on microchromosomes (13.1%) were significantly higher than were rates on macrochromosomes (10.3%; P = 0.024). Broadly assuming neutrality of introns and synonymous sites, or constraints on such sequences do not differ between chromosomal classes, these observations imply that microchromosomal genes are exposed to more germ line mutations than those on other chromosomes. We also find that dN/dS ratios for genes located on microchromosomes (average, 0.094) are significantly lower than those of macrochromosomes (average, 0.185; P = 0.025), suggesting that the proteins of genes on microchromosomes are under greater evolutionary constraint.
Figures
References
-
- Auer, H., Mayr, B., Lambrou, M., and Schleger, W. 1987. An extended chicken karyotype, including the NOR chromosome. Cytogenet. Cell Genet. 45: 218-221. - PubMed
-
- Axelsson, E., Smith, N.G.C., Sundstrom, H., Berlin, S., and Ellegren, H. 2004. Male-biased mutation rate and divergence in autosomal, Z-linked and W-linked introns of chicken and turkey. Mol. Biol. Evol. 18: 1538-1547. - PubMed
-
- Burt, D.W. 2002. Origin and evolution of avian microchromosomes. Cytogenet. Genome Res. 96: 97-112. - PubMed
-
- Burt, D.W., Bruley, C., Dunn, I.C., Jones, C.T., Ramage, A., Law, A.S., Morrice, D.R., Paton, I.R., Smith, J., Windsor, D., et al. 1999. The dynamics of chromosome evolution in birds and mammals. Nature 402: 411-413. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous
