Constitutional rearrangement of the architectural factor HMGA2: a novel human phenotype including overgrowth and lipomas
- PMID: 15593017
- PMCID: PMC1196379
- DOI: 10.1086/427565
Constitutional rearrangement of the architectural factor HMGA2: a novel human phenotype including overgrowth and lipomas
Abstract
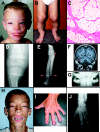
Although somatic mutations in a number of genes have been associated with development of human tumors, such as lipomas, relatively few examples exist of germline mutations in these genes. Here we describe an 8-year-old boy who has a de novo pericentric inversion of chromosome 12, with breakpoints at p11.22 and q14.3, and a phenotype including extreme somatic overgrowth, advanced endochondral bone and dental ages, a cerebellar tumor, and multiple lipomas. His chromosomal inversion was found to truncate HMGA2, a gene that encodes an architectural factor involved in the etiology of many benign mesenchymal tumors and that maps to the 12q14.3 breakpoint. Similar truncations of murine Hmga2 in transgenic mice result in somatic overgrowth and, in particular, increased abundance of fat and lipomas, features strikingly similar to those observed in the child. This represents the first report of a constitutional rearrangement affecting HMGA2 and demonstrates the role of this gene in human growth and development. Systematic genetic analysis and clinical studies of this child may offer unique insights into the role of HMGA2 in adipogenesis, osteogenesis, and general growth control.
Figures

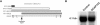
References
Electronic-Database Information
-
- Mitelman Database of Chromosome Aberrations in Cancer, http://cgap.nci.nih.gov/Chromosomes/Mitelman
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
-
- UCSC Genome Browser, May 2004 assembly, http://genome.ucsc.edu/
References
-
- Anand A, Chada K (2000) In vivo modulation of Hmgic reduces obesity. Nat Genet 24:377–380 - PubMed
-
- Andrieux J, Demory JL, Dupriez B, Quief S, Plantier I, Roumier C, Bauters F, Lai JL, Kerckaert JP (2004) Dysregulation and overexpression of HMGA2 in myelofibrosis with myeloid metaplasia. Genes Chromosomes Cancer 39:82–87 - PubMed
-
- Arlotta P, Tai AK-F, Manfioletti G, Clifford C, Jay G, Ono SJ (2000) Transgenic mice expressing a truncated form of the high mobility group I-C protein develop adiposity and an abnormally high prevalence of lipomas. J Biol Chem 275:14394–14400 - PubMed
-
- Ashar HR, Schoenberg Fejzo M, Tkachenko A, Zhou X, Fletcher JA, Weremowicz S, Morton CC, Chada K (1995) Disruption of the architectural factor HMGIC: DNA-binding AT hook motifs fused in lipomas to distinct transcriptional regulatory domains. Cell 82:57–65 - PubMed
-
- Ashar HR, Tkachenko A, Shah P, Chada K (2003) HMGA2 is expressed in an allele-specific manner in human lipomas. Cancer Genet Cytogenet 143:160–168 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

