Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome
- PMID: 15595826
- PMCID: PMC2879061
- DOI: 10.1021/bi048364h
Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome
Abstract
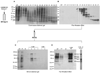
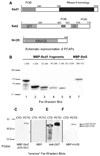
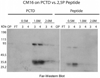
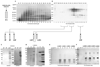
CTD kinase I (CTDK-I) of Saccharomyces cerevisiae is required for normal phosphorylation of the C-terminal repeat domain (CTD) on elongating RNA polymerase II. To elucidate cellular roles played by this kinase and the hyperphosphorylated CTD (phosphoCTD) it generates, we systematically searched yeast extracts for proteins that bound to the phosphoCTD made by CTDK-I in vitro. Initially, using a combination of far-western blotting and phosphoCTD affinity chromatography, we discovered a set of novel phosphoCTD-associating proteins (PCAPs) implicated in a variety of nuclear functions. We identified the phosphoCTD-interacting domains of a number of these PCAPs, and in several test cases (namely, Set2, Ssd1, and Hrr25) adduced evidence that phosphoCTD binding is functionally important in vivo. Employing surface plasmon resonance (BIACORE) analysis, we found that recombinant versions of these and other PCAPs bind preferentially to CTD repeat peptides carrying SerPO(4) residues at positions 2 and 5 of each seven amino acid repeat, consistent with the positional specificity of CTDK-I in vitro [Jones, J. C., et al. (2004) J. Biol. Chem. 279, 24957-24964]. Subsequently, we used a synthetic CTD peptide with three doubly phosphorylated repeats (2,5P) as an affinity matrix, greatly expanding our search for PCAPs. This resulted in identification of approximately 100 PCAPs and associated proteins representing a wide range of functions (e.g., transcription, RNA processing, chromatin structure, DNA metabolism, protein synthesis and turnover, RNA degradation, snRNA modification, and snoRNP biogenesis). The varied nature of these PCAPs and associated proteins points to an unexpectedly diverse set of connections between Pol II elongation and other processes, conceptually expanding the role played by CTD phosphorylation in functional organization of the nucleus.
Figures





References
-
- Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 1996;271:19009–19012. - PubMed
-
- Dahmus ME. Phosphorylation of the C-terminal domain of RNA polymerase II. Biochim. Biophys Acta. 1995;1261:171–182. - PubMed
-
- Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell. 2004;13:67–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

