Genome evolution in the genus Sorghum (Poaceae)
- PMID: 15596469
- PMCID: PMC4246720
- DOI: 10.1093/aob/mci015
Genome evolution in the genus Sorghum (Poaceae)
Abstract
Background and aims: The roles of variation in DNA content in plant evolution and adaptation remain a major biological enigma. Chromosome number and 2C DNA content were determined for 21 of the 25 species of the genus Sorghum and analysed from a phylogenetic perspective.
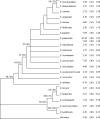
Methods: DNA content was determined by flow cytometry. A Sorghum phylogeny was constructed based on combined nuclear ITS and chloroplast ndhF DNA sequences.
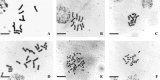
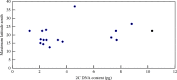
Key results: Chromosome counts (2n = 10, 20, 30, 40) were, with few exceptions, concordant with published numbers. New chromosome numbers were obtained for S. amplum (2n = 30) and S. leiocladum (2n = 10). 2C DNA content varies 8.1-fold (1.27-10.30 pg) among the 21 Sorghum species. 2C DNA content varies 3.6-fold from 1.27 pg to 4.60 pg among the 2n = 10 species and 5.8-fold (1.52-8.79 pg) among the 2n = 20 species. The x = 5 genome size varies over an 8.8-fold range from 0.26 pg to 2.30 pg. The mean 2C DNA content of perennial species (6.20 pg) is significantly greater than the mean (2.92 pg) of the annuals. Among the 21 species studied, the mean x = 5 genome size of annuals (1.15 pg) and of perennials (1.29 pg) is not significantly different. Statistical analysis of Australian species showed: (a) mean 2C DNA content of annual (2.89 pg) and perennial (7.73 pg) species is significantly different; (b) mean x = 5 genome size of perennials (1.66 pg) is significantly greater than that of the annuals (1.09 pg); (c) the mean maximum latitude at which perennial species grow (-25.4 degrees) is significantly greater than the mean maximum latitude (-17.6) at which annual species grow.
Conclusions: The DNA sequence phylogeny splits Sorghum into two lineages, one comprising the 2n = 10 species with large genomes and their polyploid relatives, and the other with the 2n = 20, 40 species with relatively small genomes. An apparent phylogenetic reduction in genome size has occurred in the 2n = 10 lineage. Genome size evolution in the genus Sorghum apparently did not involve a 'one way ticket to genomic obesity' as has been proposed for the grasses.
Figures
Similar articles
-
[Molecular phylogenetic relationships among species in the genus Sorghum based on partial Adh1 gene].Yi Chuan. 2009 May;31(5):523-30. doi: 10.3724/sp.j.1005.2009.00523. Yi Chuan. 2009. PMID: 19586848 Chinese.
-
Genome size variation in Central European species of Cirsium (Compositae) and their natural hybrids.Ann Bot. 2004 Sep;94(3):353-63. doi: 10.1093/aob/mch151. Epub 2004 Aug 3. Ann Bot. 2004. PMID: 15292040 Free PMC article.
-
Nuclear DNA assay in solving issues related to ancestry of the domesticated diploid safflower (Carthamus tinctorius L.) and the polyploid (Carthamus) taxa, and phylogenetic and genomic relationships in the genus Carthamus L. (Asteraceae).Mol Phylogenet Evol. 2009 Dec;53(3):631-44. doi: 10.1016/j.ympev.2009.07.012. Epub 2009 Jul 12. Mol Phylogenet Evol. 2009. PMID: 19602441
-
[Trends of angiosperm genome evolution].Tsitologiia. 2011;53(4):295-312. Tsitologiia. 2011. PMID: 21675209 Review. Russian.
-
The origin, evolution and proposed stabilization of the terms 'genome size' and 'C-value' to describe nuclear DNA contents.Ann Bot. 2005 Jan;95(1):255-60. doi: 10.1093/aob/mci019. Ann Bot. 2005. PMID: 15596473 Free PMC article. Review.
Cited by
-
Genome-wide association analysis of the strength of the MAMP-elicited defense response and resistance to target leaf spot in sorghum.Sci Rep. 2020 Nov 30;10(1):20817. doi: 10.1038/s41598-020-77684-w. Sci Rep. 2020. PMID: 33257818 Free PMC article.
-
Exploring giant plant genomes with next-generation sequencing technology.Chromosome Res. 2011 Oct;19(7):939-53. doi: 10.1007/s10577-011-9246-z. Chromosome Res. 2011. PMID: 21987187 Review.
-
Infrageneric phylogeny and temporal divergence of Sorghum (Andropogoneae, Poaceae) based on low-copy nuclear and plastid sequences.PLoS One. 2014 Aug 14;9(8):e104933. doi: 10.1371/journal.pone.0104933. eCollection 2014. PLoS One. 2014. PMID: 25122516 Free PMC article.
-
Phylogenetic reconstruction using four low-copy nuclear loci strongly supports a polyphyletic origin of the genus Sorghum.Ann Bot. 2015 Aug;116(2):291-9. doi: 10.1093/aob/mcv097. Epub 2015 Jul 2. Ann Bot. 2015. PMID: 26141132 Free PMC article.
-
Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion.Ann Bot. 2005 Jan;95(1):147-75. doi: 10.1093/aob/mci010. Ann Bot. 2005. PMID: 15596464 Free PMC article. Review.
References
-
- Bennett MD. 1972. Nuclear DNA content and minimum generation time in herbaceous angiosperms. Proceedings of the Royal Society of London, Series B 181: 109–135. - PubMed
-
- Bennett MD. 1973. Nuclear characters in plants. Brookhaven Symposium in Biology 25: 344–366.
-
- Bennett MD. 1987. Variation in genomic form in plants and its ecological implications. New Phytologist 106 (Suppl.): 177–200.
-
- Bennett MD, Bhandol P, Leitch I.J. 2000. Nuclear DNA amounts in angiosperms and their modern uses—807 new estimates. Annals of Botany 86: 859–909.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

