Functional roles of nonconserved structural segments in CFTR's NH2-terminal nucleotide binding domain
- PMID: 15596536
- PMCID: PMC2217481
- DOI: 10.1085/jgp.200409174
Functional roles of nonconserved structural segments in CFTR's NH2-terminal nucleotide binding domain
Abstract
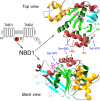
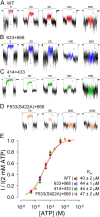
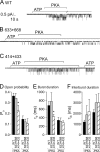
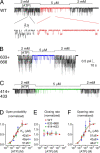
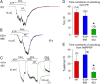
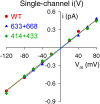
The cystic fibrosis transmembrane conductance regulator (CFTR), encoded by the gene mutated in cystic fibrosis patients, belongs to the family of ATP-binding cassette (ABC) proteins, but, unlike other members, functions as a chloride channel. CFTR is activated by protein kinase A (PKA)-mediated phosphorylation of multiple sites in its regulatory domain, and gated by binding and hydrolysis of ATP at its two nucleotide binding domains (NBD1, NBD2). The recent crystal structure of NBD1 from mouse CFTR (Lewis, H.A., S.G. Buchanan, S.K. Burley, K. Conners, M. Dickey, M. Dorwart, R. Fowler, X. Gao, W.B. Guggino, W.A. Hendrickson, et al. 2004. EMBO J. 23:282-293) identified two regions absent from structures of all other NBDs determined so far, a "regulatory insertion" (residues 404-435) and a "regulatory extension" (residues 639-670), both positioned to impede formation of the putative NBD1-NBD2 dimer anticipated to occur during channel gating; as both segments appeared highly mobile and both contained consensus PKA sites (serine 422, and serines 660 and 670, respectively), it was suggested that their phosphorylation-linked conformational changes might underlie CFTR channel regulation. To test that suggestion, we coexpressed in Xenopus oocytes CFTR residues 1-414 with residues 433-1480, or residues 1-633 with 668-1480, to yield split CFTR channels (called 414+433 and 633+668) that lack most of the insertion, or extension, respectively. In excised patches, regulation of the resulting CFTR channels by PKA and by ATP was largely normal. Both 414+433 channels and 633+668 channels, as well as 633(S422A)+668 channels (lacking both the extension and the sole PKA consensus site in the insertion), were all shut during exposure to MgATP before addition of PKA, but activated like wild type (WT) upon phosphorylation; this indicates that inhibitory regulation of nonphosphorylated WT channels depends upon neither segment. Detailed kinetic analysis of 414+433 channels revealed intact ATP dependence of single-channel gating kinetics, but slightly shortened open bursts and faster closing from the locked-open state (elicited by ATP plus pyrophosphate or ATP plus AMPPNP). In contrast, 633+668 channel function was indistinguishable from WT at both macroscopic and microscopic levels. We conclude that neither nonconserved segment is an essential element of PKA- or nucleotide-dependent regulation.
Figures
Similar articles
-
Prolonged nonhydrolytic interaction of nucleotide with CFTR's NH2-terminal nucleotide binding domain and its role in channel gating.J Gen Physiol. 2003 Sep;122(3):333-48. doi: 10.1085/jgp.200308798. J Gen Physiol. 2003. PMID: 12939393 Free PMC article.
-
The two ATP binding sites of cystic fibrosis transmembrane conductance regulator (CFTR) play distinct roles in gating kinetics and energetics.J Gen Physiol. 2006 Oct;128(4):413-22. doi: 10.1085/jgp.200609622. Epub 2006 Sep 11. J Gen Physiol. 2006. PMID: 16966475 Free PMC article.
-
Severed molecules functionally define the boundaries of the cystic fibrosis transmembrane conductance regulator's NH(2)-terminal nucleotide binding domain.J Gen Physiol. 2000 Aug;116(2):163-80. doi: 10.1085/jgp.116.2.163. J Gen Physiol. 2000. PMID: 10919864 Free PMC article.
-
ATP hydrolysis cycles and the gating of CFTR Cl- channels.Acta Physiol Scand Suppl. 1998 Aug;643:247-56. Acta Physiol Scand Suppl. 1998. PMID: 9789567 Review.
-
Control of CFTR channel gating by phosphorylation and nucleotide hydrolysis.Physiol Rev. 1999 Jan;79(1 Suppl):S77-S107. doi: 10.1152/physrev.1999.79.1.S77. Physiol Rev. 1999. PMID: 9922377 Review.
Cited by
-
Gating of the CFTR Cl- channel by ATP-driven nucleotide-binding domain dimerisation.J Physiol. 2009 May 15;587(Pt 10):2151-61. doi: 10.1113/jphysiol.2009.171595. Epub 2009 Mar 30. J Physiol. 2009. PMID: 19332488 Free PMC article. Review.
-
Regulatory insertion removal restores maturation, stability and function of DeltaF508 CFTR.J Mol Biol. 2010 Aug 13;401(2):194-210. doi: 10.1016/j.jmb.2010.06.019. Epub 2010 Jun 16. J Mol Biol. 2010. PMID: 20561529 Free PMC article.
-
Jump into a New Fold-A Homology Based Model for the ABCG2/BCRP Multidrug Transporter.PLoS One. 2016 Oct 14;11(10):e0164426. doi: 10.1371/journal.pone.0164426. eCollection 2016. PLoS One. 2016. PMID: 27741279 Free PMC article.
-
Bioelectric regulation of intestinal stem cells.Trends Cell Biol. 2023 Jul;33(7):555-567. doi: 10.1016/j.tcb.2022.10.003. Epub 2022 Nov 15. Trends Cell Biol. 2023. PMID: 36396487 Free PMC article. Review.
-
Conformational changes in the catalytically inactive nucleotide-binding site of CFTR.J Gen Physiol. 2013 Jul;142(1):61-73. doi: 10.1085/jgp.201210954. Epub 2013 Jun 10. J Gen Physiol. 2013. PMID: 23752332 Free PMC article.
References
-
- Aleksandrov, L., A. Mengos, X. Chang, A. Aleksandrov, and J.R. Riordan. 2001. Differential interactions of nucleotides at the two nucleotide binding domains of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 276:12918–12923. - PubMed
-
- Aleksandrov, L., A.A. Aleksandrov, X.B. Chang, and J.R. Riordan. 2002. The first nucleotide binding domain of cystic fibrosis transmembrane conductance regulator is a site of stable nucleotide interaction, whereas the second is a site of rapid turnover. J. Biol. Chem. 277:15419–15425. - PubMed
-
- Anderson, M.P., and M.J. Welsh. 1992. Regulation by ATP and ADP of CFTR chloride channels that contain mutant nucleotide-binding domains. Science. 257:1701–1704. - PubMed
-
- Anderson, M.P., H.A. Berger, D.P. Rich, R.J. Gregory, A.E. Smith, and M.J. Welsh. 1991. Nucleoside triphosphates are required to open the CFTR chloride channel. Cell. 67:775–784. - PubMed
-
- Armstrong, S., L. Tabernero, H. Zhang, M. Hermodson, and C. Stauffacher. 1998. The 2.5 Å structure of the N-terminal ATP-binding cassette of the ribose ABC transporter. Biophys. J. 74:A338.

