Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression
- PMID: 15596808
- PMCID: PMC538702
- DOI: 10.1128/JVI.79.1.124-131.2005
Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression
Abstract
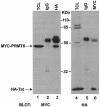
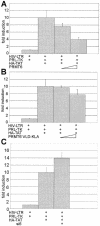
The human immunodeficiency virus (HIV) transactivator protein, Tat, stimulates transcription from the viral long terminal repeats via an arginine-rich transactivating domain. Since arginines are often known to be methylated, we investigated whether HIV type 1 (HIV-1) Tat was a substrate for known protein arginine methyltransferases (PRMTs). Here we identify Tat as a substrate for the arginine methyltransferase, PRMT6. Tat is specifically associated with and methylated by PRMT6 within cells. Overexpression of wild-type PRMT6, but not a methylase-inactive PRMT6 mutant, decreased Tat transactivation of an HIV-1 long terminal repeat luciferase reporter plasmid in a dose-dependent manner. Knocking down PRMT6 consistently increased HIV-1 production in HEK293T cells and also led to increased viral infectiousness as shown in multinuclear activation of a galactosidase indicator assays. Our study demonstrates that arginine methylation of Tat negatively regulates its transactivation activity and that PRMT6 acts as a restriction factor for HIV replication.
Figures



Similar articles
-
Arginine methylation of the human immunodeficiency virus type 1 Tat protein by PRMT6 negatively affects Tat Interactions with both cyclin T1 and the Tat transactivation region.J Virol. 2007 Apr;81(8):4226-34. doi: 10.1128/JVI.01888-06. Epub 2007 Jan 31. J Virol. 2007. PMID: 17267505 Free PMC article.
-
PRMT6 diminishes HIV-1 Rev binding to and export of viral RNA.Retrovirology. 2006 Dec 18;3:93. doi: 10.1186/1742-4690-3-93. Retrovirology. 2006. PMID: 17176473 Free PMC article.
-
The protein arginine methyltransferase PRMT6 inhibits HIV-1 Tat nucleolar retention.Biochim Biophys Acta. 2016 Feb;1863(2):254-62. doi: 10.1016/j.bbamcr.2015.11.019. Epub 2015 Dec 2. Biochim Biophys Acta. 2016. PMID: 26611710
-
Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator.IUBMB Life. 2001 Mar;51(3):175-81. doi: 10.1080/152165401753544241. IUBMB Life. 2001. PMID: 11547919 Review.
-
Will diverse Tat interactions lead to novel antiretroviral drug targets?Curr Drug Targets. 2006 Dec;7(12):1595-606. doi: 10.2174/138945006779025338. Curr Drug Targets. 2006. PMID: 17168834 Review.
Cited by
-
Expression profiling of chromatin-modifying enzymes and global DNA methylation in CD4+ T cells from patients with chronic HIV infection at different HIV control and progression states.Clin Epigenetics. 2018 Feb 13;10:20. doi: 10.1186/s13148-018-0448-5. eCollection 2018. Clin Epigenetics. 2018. PMID: 29449904 Free PMC article.
-
Phosphorylation of HIV-1 Tat by CDK2 in HIV-1 transcription.Retrovirology. 2006 Nov 3;3:78. doi: 10.1186/1742-4690-3-78. Retrovirology. 2006. PMID: 17083724 Free PMC article.
-
The Cellular lysine methyltransferase Set7/9-KMT7 binds HIV-1 TAR RNA, monomethylates the viral transactivator Tat, and enhances HIV transcription.Cell Host Microbe. 2010 Mar 18;7(3):234-44. doi: 10.1016/j.chom.2010.02.005. Cell Host Microbe. 2010. PMID: 20227666 Free PMC article.
-
OsPRMT6b balances plant growth and high temperature stress by feedback inhibition of abscisic acid signaling.Nat Commun. 2025 Jun 4;16(1):5173. doi: 10.1038/s41467-025-60350-y. Nat Commun. 2025. PMID: 40467572 Free PMC article.
-
PRMT2 promotes HIV-1 latency by preventing nucleolar exit and phase separation of Tat into the Super Elongation Complex.Nat Commun. 2023 Nov 10;14(1):7274. doi: 10.1038/s41467-023-43060-1. Nat Commun. 2023. PMID: 37949879 Free PMC article.
References
-
- An, W., J. Kim, and R. G. Roeder. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117:735-748. - PubMed
-
- Bedford, M. T., A. Frankel, M. B. Yaffe, S. Clarke, P. Leder, and S. Richard. 2000. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem. 275:16030-16036. - PubMed
-
- Boisvert, F. M., J. Cote, M. C. Boulanger, and S. Richard. 2003. A proteomic analysis of arginine-methylated protein complexes. Mol. Cell Proteomics 2:1319-1330. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

